Diverse phage-encoded toxins in a protective insect endosymbiont - PubMed (original) (raw)
Diverse phage-encoded toxins in a protective insect endosymbiont
Patrick H Degnan et al. Appl Environ Microbiol. 2008 Nov.
Abstract
The lysogenic bacteriophage APSE infects "Candidatus Hamiltonella defensa," a facultative endosymbiont of aphids and other sap-feeding insects. This endosymbiont has established a beneficial association with aphids, increasing survivorship following attack by parasitoid wasps. Although APSE and "Ca. Hamiltonella defensa" are effectively maternally transmitted between aphid generations, they can also be horizontally transferred among insect hosts, which results in genetically distinct "Ca. Hamiltonella defensa" strains infecting the same aphid species and sporadic distributions of both APSE and "Ca. Hamiltonella defensa" among hosts. Aphids infected only with "Ca. Hamiltonella defensa" have significantly less protection than those infected with both "Ca. Hamiltonella defensa" and APSE. This protection has been proposed to be connected to eukaryote-targeted toxins previously discovered in the genomes of two characterized APSE strains. In this study, we have sequenced partial genomes from seven additional APSE strains to address the evolution and extent of toxin variation in this phage. The APSE lysis region has been a hot spot for nonhomologous recombination of novel virulence cassettes. We identified four new toxins from three protein families, Shiga-like toxin, cytolethal distending toxin, and YD-repeat toxins. These recombination events have also resulted in reassortment of the downstream lysozyme and holin genes. Analysis of the conserved APSE genes flanking the variable toxin cassettes reveals a close phylogenetic association with phage sequences from two other facultative endosymbionts of insects. Thus, phage may act as a conduit for ongoing gene exchange among heritable endosymbionts.
Figures
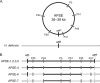
FIG. 1.
APSE genome amplification by PCR. (A) Schematic diagram of the circularly permutated APSE chromosome, where gray bars indicate the six anchor sequences used to amplify genomic fragments. The APSE integration site in the “Ca. Hamiltonella defensa” chromosome was determined from the draft “Ca. Hamiltonella defensa” strain 5AT genome (Degnan and Moran, unpublished). The attachment site in APSE (attP) is adjacent to the integrase gene P38 and corresponds to an identical 62-bp sequence in “Ca. Hamiltonella defensa” (attB), which overlaps with an arginine tRNA. (B) All six of the genomic fragments were amplified for the novel strains APSE-3, -4, and -5 (solid black lines) and for the previously sequenced strains APSE-1 and -2. Failure to amplify several interanchor fragments from APSE-6 and -7 (dashed lines) suggests recombination or degradation of the phage. One or both of the APSE/“Ca. Hamiltonella defensa” junctures have been confirmed by PCR for APSE-1, -2, -3, -4, -5, and -6 (vertical black bars).
FIG. 2.
Variable-length regions of the APSE chromosomes. (A) Map of the entire APSE-1 chromosome. Boxes above the line represent ORFs transcribed in the rightward direction; below the line, leftward. The six anchor genes are indicated, and the dashed boxes show the two regions found to be variable in length by PCR. (B and C) Detailed view of the sequenced virulence cassette (B) or DNA replication regions (C). Orthologous ORFs are shown in the same color, and putatively toxic ORFs are shown with hatch marks. Putative pseudogenes (ψ) are shown as boxes bordered with dashed lines.
FIG. 3.
Amino acid phylogenies of APSE-encoded toxins. Consensus maximum likelihood phylogenies of APSE-encoded cytolethal distending toxin (CdtB) (A) or YD-repeat proteins (B). Proteins previously demonstrated to be toxic are indicated with daggers (†). Bootstrap values from PhyML, followed by posterior probabilities estimated in MrBayes, are listed at appropriate nodes. Nodes with less than 50% support by both methods have been collapsed. Genus abbreviations are as follows: H., Haemophilus (for H. ducreyi) or Helicobacter (all others); C., Campylobacter; F., “_Flexispira_”; S., Shigella (for S. boydii) or Salmonella (for S. enterica); P., Photorhabdus (for P. luminescens) or Pseudomonas (all others); A., Aggregatibacter. sv., serovar; str., strain.
FIG. 4.
APSE lysozyme protein phylogeny. The consensus maximum-likelihood phylogeny for the APSE lysozymes P13 and “F” is shown. These lysozymes form two distinct clades, each of which is most closely related to homologs found in Sodalis glossinidius, the facultative endosymbiont of tsetse flies (Glossinia morsitans). The brackets denote that P13-like lysozymes are all linked to an adjacent group I holin while F-like lysozymes are linked to a group II holin. Numbers in parentheses correspond to the gene number from the genome sequenced, and the asterisks indicate the two APSE strains that carry both a group I and a group II holin gene. The support values at the nodes list the bootstrap values from PhyML followed by posterior probabilities estimated in MrBayes. Genera are as follows: Y., Yersinia; G., Glossinia; V., Vibrio; S., Sodalis.
Similar articles
- Evolutionary genetics of a defensive facultative symbiont of insects: exchange of toxin-encoding bacteriophage.
Degnan PH, Moran NA. Degnan PH, et al. Mol Ecol. 2008 Feb;17(3):916-29. doi: 10.1111/j.1365-294X.2007.03616.x. Epub 2007 Dec 20. Mol Ecol. 2008. PMID: 18179430 - The Protector within: Comparative Genomics of APSE Phages across Aphids Reveals Rampant Recombination and Diverse Toxin Arsenals.
Rouïl J, Jousselin E, Coeur d'acier A, Cruaud C, Manzano-Marín A. Rouïl J, et al. Genome Biol Evol. 2020 Jun 1;12(6):878-889. doi: 10.1093/gbe/evaa089. Genome Biol Evol. 2020. PMID: 32386316 Free PMC article. - Arsenophonus insect symbionts are commonly infected with APSE, a bacteriophage involved in protective symbiosis.
Duron O. Duron O. FEMS Microbiol Ecol. 2014 Oct;90(1):184-94. doi: 10.1111/1574-6941.12381. Epub 2014 Aug 11. FEMS Microbiol Ecol. 2014. PMID: 25041857 - Variations on a protective theme: Hamiltonella defensa infections in aphids variably impact parasitoid success.
Oliver KM, Higashi CH. Oliver KM, et al. Curr Opin Insect Sci. 2019 Apr;32:1-7. doi: 10.1016/j.cois.2018.08.009. Epub 2018 Aug 30. Curr Opin Insect Sci. 2019. PMID: 31113620 Review. - Facultative symbionts in aphids and the horizontal transfer of ecologically important traits.
Oliver KM, Degnan PH, Burke GR, Moran NA. Oliver KM, et al. Annu Rev Entomol. 2010;55:247-66. doi: 10.1146/annurev-ento-112408-085305. Annu Rev Entomol. 2010. PMID: 19728837 Review.
Cited by
- A facultative endosymbiont in aphids can provide diverse ecological benefits.
Heyworth ER, Ferrari J. Heyworth ER, et al. J Evol Biol. 2015 Oct;28(10):1753-60. doi: 10.1111/jeb.12705. Epub 2015 Aug 18. J Evol Biol. 2015. PMID: 26206380 Free PMC article. - Bacterial communities in natural versus pesticide-treated Aphis gossypii populations in North China.
Zhang S, Luo J, Wang L, Zhang L, Zhu X, Jiang W, Cui J. Zhang S, et al. Microbiologyopen. 2019 Mar;8(3):e00652. doi: 10.1002/mbo3.652. Epub 2018 Jun 7. Microbiologyopen. 2019. PMID: 29877631 Free PMC article. - The price of protection: a defensive endosymbiont impairs nymph growth in the bird cherry-oat aphid, Rhopalosiphum padi.
Leybourne DJ, Bos JIB, Valentine TA, Karley AJ. Leybourne DJ, et al. Insect Sci. 2020 Feb;27(1):69-85. doi: 10.1111/1744-7917.12606. Epub 2018 Jul 25. Insect Sci. 2020. PMID: 29797656 Free PMC article. - The toxins of vertically transmitted Spiroplasma.
Moore LD, Ballinger MJ. Moore LD, et al. Front Microbiol. 2023 May 18;14:1148263. doi: 10.3389/fmicb.2023.1148263. eCollection 2023. Front Microbiol. 2023. PMID: 37275155 Free PMC article.
References
- AbuOun, M., G. Manning, S. A. Cawthraw, A. Ridley, I. H. Ahmed, T. M. Wassenaar, and D. G. Newell. 2005. Cytolethal distending toxin (CDT)-negative Campylobacter jejuni strains and anti-CDT neutralizing antibodies are induced during human infection but not during colonization in chickens. Infect. Immun. 73:3053-3062. - PMC - PubMed
- Akman, L., A. Yamashita, H. Watanabe, K. Oshima, T. Shiba, M. Hattori, and S. Aksoy. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402-407. - PubMed
- Allen, M. J., G. F. White, and A. P. Morby. 2006. The response of Escherichia coli to exposure to the biocide polyhexamethylene biguanide. Microbiology 152:989-1000. - PubMed
- Altschul, S. F., W. Gish, W. Miller, W. E. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous