Convergent evolution in the genetic basis of Müllerian mimicry in heliconius butterflies - PubMed (original) (raw)
Convergent evolution in the genetic basis of Müllerian mimicry in heliconius butterflies
Simon W Baxter et al. Genetics. 2008 Nov.
Abstract
The neotropical butterflies Heliconius melpomene and H. erato are Müllerian mimics that display the same warningly colored wing patterns in local populations, yet pattern diversity between geographic regions. Linkage mapping has previously shown convergent red wing phenotypes in these species are controlled by loci on homologous chromosomes. Here, AFLP bulk segregant analysis using H. melpomene crosses identified genetic markers tightly linked to two red wing-patterning loci. These markers were used to screen a H. melpomene BAC library and a tile path was assembled spanning one locus completely and part of the second. Concurrently, a similar strategy was used to identify a BAC clone tightly linked to the locus controlling the mimetic red wing phenotypes in H. erato. A methionine rich storage protein (MRSP) gene was identified within this BAC clone, and comparative genetic mapping shows red wing color loci are in homologous regions of the genome of H. erato and H. melpomene. Subtle differences in these convergent phenotypes imply they evolved independently using somewhat different developmental routes, but are nonetheless regulated by the same switch locus. Genetic mapping of MRSP in a third related species, the "tiger" patterned H. numata, has no association with wing patterning and shows no evidence for genomic translocation of wing-patterning loci.
Figures
Figure 1.—
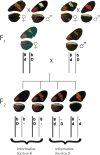
Heliconius melpomene and H. erato comimics. D phenotype: Dorsal and ventral wings from H. melpomene ecuadorensis and H. erato etylus, displaying the D phenotype. Specimens were collected near Zamora, Ecuador. The yellow forewing patch is controlled by HeD in H. erato and by a second locus (called N) in H. melpomene. B phenotype: H. melpomene melpomene and H. erato hydara were collected near Cayenne, French Guiana (4°91′ N, 52°36′ W). All individuals are males.
Figure 2.—
HmB and HmD phenotypes are inherited in repulsion. Schematic of LG18 chromosomes from an F1 cross between the heterozygous Bb/Dd female (dark gray) and male (light gray). Crossing over of sister chromatids does not occur during oogenesis, so F2 progeny inherit either bD or Bd genotypes from the F1 female. As all LG18 chromosomal markers are completely linked when inherited from the F1 mother, molecular markers are used to assign chromosome prints. Recombination between HmD and HmB or Hmd and Hmb may occur in the F1 male during spermatogenesis. Consequently, phenotypes are only informative for one of the B/b alleles or one of the D/d alleles and are therefore in repulsion. For example in the case of a bD/b- F2 individual, the F1 female-derived chromosome contributed b/D and F1 male-derived chromosome b/-. Here, it is unclear whether one or two HmD alleles are present, as the F1 female HmD phenotype masks any contribution by the F1 male. Male informative genotypes from heterozygous B-/D- progeny were deduced using chromosome prints. A B-/D- F2 heterozygote that inherited a b/D chromosome from the F1 female must have inherited a dominant HmB allele from the F1 father to produce the observed phenotype. A hyphen “-” signifies unknown male-derived alleles.
Figure 3.—
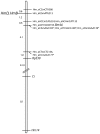
Linkage map of H. melpomene LG18 (77.3 cM, log likelihood = −92.61). Three nuclear genes (Ci, RpS30, and Bm44), one microsatellite marker (Hm14), and six AFLP sequence-tagged sites were genetically mapped in 164 F2 individuals from brood 44. The linkage map was constructed using MapMaker 2.0. Four AFLP markers identified by an asterisk were positioned manually on the basis of AFLP banding patterns for up to 35 individuals. All AFLP markers are within a 6.5-cM region.
Figure 4.—
Analysis of recombinants spanning the HmB/D region on LG18. A H. melpomene BAC library was probed with three AFLP markers (open circles) linked to the HmB and HmD loci. Hm_eACTmGTA195 hybridized with 1 BAC clone, bHM40C14, within contig 1729. Hm_eCCmCTA386 hybridized to 19 clones from contig 196, (containing 32 clones). Hm_eCGmATG213 hybridized to 6 clones from contig 446 (containing 6 clones) plus 5 clones from contig 936 (containing 10 clones). PCR amplification using BAC end primers designed from contig 446 successfully amplified in BAC clones from contig 936, and vice versa, demonstrating these two contigs overlap (supplemental Figure 1). This implies contigs 446 and 936 may represent very divergent haplotypes for the B/D region, as fingerprinting did not group these clones into a single contig. The BAC ends of some clones were sequenced and used to design genotyping assays for the 372 progeny (solid circles). As HmB and HmD are inherited in repulsion, recombinants from BAC ends to these loci are listed independently. Recombinants between contig ends were observed, enabling orientation of the HmB and HmD loci. The H. melpomene BAC library was screened using additional BAC end probes (solid squares), bHM40A21_sp6 (contig 936), bHM28L23_sp6 (contig 1305), and bHM36K2_sp6 (contig 196), which created a single tile path spanning the HmB locus and part of the HmD locus. Finally, a gene near the H. erato HeD locus (MRSP) was used as a probe. For full details of BAC screening results see supplemental Table 1. Accession nos. for BAC end sequences are FI107283–FI107295 and FI136209–FI136223. The figure scale is based on relative sizes of fingerprinted contigs.
Figure 5.—
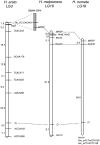
Linkage maps comparing homologous chromosomes from H. erato, H. melpomene, and H. numata. H. erato linkage map adapted from K
apan
et al. (2006), showing HeD in the most likely position (70% probability density). BAC library screening using H. erato AFLP marker He_CCAC491 identified clone BBAM-25K4. Sequence analysis identified a MRSP gene, which was genetically mapped along with Ci in all three species. Two H. melpomene AFLP markers (Hm_eACTmGTA195 and Hm_eCTmCTC156) and one microsatellite (Hm14) cross amplified in H. numata and were genetically mapped to LG18. Numbers shown are centimorgans.
Similar articles
- Highly conserved gene order and numerous novel repetitive elements in genomic regions linked to wing pattern variation in Heliconius butterflies.
Papa R, Morrison CM, Walters JR, Counterman BA, Chen R, Halder G, Ferguson L, Chamberlain N, Ffrench-Constant R, Kapan DD, Jiggins CD, Reed RD, McMillan WO. Papa R, et al. BMC Genomics. 2008 Jul 22;9:345. doi: 10.1186/1471-2164-9-345. BMC Genomics. 2008. PMID: 18647405 Free PMC article. - Convergent, modular expression of ebony and tan in the mimetic wing patterns of Heliconius butterflies.
Ferguson LC, Maroja L, Jiggins CD. Ferguson LC, et al. Dev Genes Evol. 2011 Dec;221(5-6):297-308. doi: 10.1007/s00427-011-0380-6. Epub 2011 Dec 3. Dev Genes Evol. 2011. PMID: 22139062 - Comparative population genetics of a mimicry locus among hybridizing Heliconius butterfly species.
Chamberlain NL, Hill RI, Baxter SW, Jiggins CD, Kronforst MR. Chamberlain NL, et al. Heredity (Edinb). 2011 Sep;107(3):200-4. doi: 10.1038/hdy.2011.3. Epub 2011 Feb 9. Heredity (Edinb). 2011. PMID: 21304546 Free PMC article. - The 2023 Latin America report of the Lancet Countdown on health and climate change: the imperative for health-centred climate-resilient development.
Hartinger SM, Palmeiro-Silva YK, Llerena-Cayo C, Blanco-Villafuerte L, Escobar LE, Diaz A, Sarmiento JH, Lescano AG, Melo O, Rojas-Rueda D, Takahashi B, Callaghan M, Chesini F, Dasgupta S, Posse CG, Gouveia N, Martins de Carvalho A, Miranda-Chacón Z, Mohajeri N, Pantoja C, Robinson EJZ, Salas MF, Santiago R, Sauma E, Santos-Vega M, Scamman D, Sergeeva M, Souza de Camargo T, Sorensen C, Umaña JD, Yglesias-González M, Walawender M, Buss D, Romanello M. Hartinger SM, et al. Lancet Reg Health Am. 2024 Apr 23;33:100746. doi: 10.1016/j.lana.2024.100746. eCollection 2024 May. Lancet Reg Health Am. 2024. PMID: 38800647 Free PMC article. Review. - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
Cited by
- Hybrid speciation in Heliconius butterflies? A review and critique of the evidence.
Brower AV. Brower AV. Genetica. 2011 May;139(5):589-609. doi: 10.1007/s10709-010-9530-4. Epub 2010 Nov 28. Genetica. 2011. PMID: 21113790 Free PMC article. Review. - Haplotype tagging reveals parallel formation of hybrid races in two butterfly species.
Meier JI, Salazar PA, Kučka M, Davies RW, Dréau A, Aldás I, Box Power O, Nadeau NJ, Bridle JR, Rolian C, Barton NH, McMillan WO, Jiggins CD, Chan YF. Meier JI, et al. Proc Natl Acad Sci U S A. 2021 Jun 22;118(25):e2015005118. doi: 10.1073/pnas.2015005118. Proc Natl Acad Sci U S A. 2021. PMID: 34155138 Free PMC article. - Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing.
Nadeau NJ, Whibley A, Jones RT, Davey JW, Dasmahapatra KK, Baxter SW, Quail MA, Joron M, ffrench-Constant RH, Blaxter ML, Mallet J, Jiggins CD. Nadeau NJ, et al. Philos Trans R Soc Lond B Biol Sci. 2012 Feb 5;367(1587):343-53. doi: 10.1098/rstb.2011.0198. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22201164 Free PMC article. - The genetic architecture of adaptation: convergence and pleiotropy in Heliconius wing pattern evolution.
Morris J, Navarro N, Rastas P, Rawlins LD, Sammy J, Mallet J, Dasmahapatra KK. Morris J, et al. Heredity (Edinb). 2019 Aug;123(2):138-152. doi: 10.1038/s41437-018-0180-0. Epub 2019 Jan 22. Heredity (Edinb). 2019. PMID: 30670842 Free PMC article. - Genome-wide introgression among distantly related Heliconius butterfly species.
Zhang W, Dasmahapatra KK, Mallet J, Moreira GR, Kronforst MR. Zhang W, et al. Genome Biol. 2016 Feb 27;17:25. doi: 10.1186/s13059-016-0889-0. Genome Biol. 2016. PMID: 26921238 Free PMC article.
References
- Brugmans, B., A. Fernandez del Carmen, C. W. B. Bachem, H. van Os, H. van Eck et al., 2002. A novel method for the construction of genome wide transcriptome maps. Plant J. 31 211–222. - PubMed
- Charlesworth, D., and B. Charlesworth, 1975. Theoretical genetics of Batesian mimicry. 2. Evolution of supergenes. J. Theor. Biol. 55 305–324. - PubMed
- Colosimo, P., K. Hosemann, S. Balabhadra, G. J. Villarreal, M. Dickson et al., 2005. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307 1928–1933. - PubMed
- Gahan, L. J., F. Gould and D. G. Heckel, 2001. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 293 857–860. - PubMed
- Heckel, D. G., L. J. Gahan, J. C. Daly and S. Trowell, 1998. A genomic approach to understanding Heliothis and Helicoverpa resistance to chemical and biological insecticides. Philos. Trans. R. Soc. Lond. B, Biol. Sci. 353 1713–1722.
Publication types
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- BB/E008836/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E011845/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources