Modeling spinal muscular atrophy in Drosophila - PubMed (original) (raw)
Modeling spinal muscular atrophy in Drosophila
Howard Chia-Hao Chang et al. PLoS One. 2008.
Abstract
Spinal Muscular Atrophy (SMA), a recessive hereditary neurodegenerative disease in humans, has been linked to mutations in the survival motor neuron (SMN) gene. SMA patients display early onset lethality coupled with motor neuron loss and skeletal muscle atrophy. We used Drosophila, which encodes a single SMN ortholog, survival motor neuron (Smn), to model SMA, since reduction of Smn function leads to defects that mimic the SMA pathology in humans. Here we show that a normal neuromuscular junction (NMJ) structure depends on SMN expression and that SMN concentrates in the post-synaptic NMJ regions. We conducted a screen for genetic modifiers of an Smn phenotype using the Exelixis collection of transposon-induced mutations, which affects approximately 50% of the Drosophila genome. This screen resulted in the recovery of 27 modifiers, thereby expanding the genetic circuitry of Smn to include several genes not previously known to be associated with this locus. Among the identified modifiers was wishful thinking (wit), a type II BMP receptor, which was shown to alter the Smn NMJ phenotype. Further characterization of two additional members of the BMP signaling pathway, Mothers against dpp (Mad) and Daughters against dpp (Dad), also modify the Smn NMJ phenotype. The NMJ defects caused by loss of Smn function can be ameliorated by increasing BMP signals, suggesting that increased BMP activity in SMA patients may help to alleviate symptoms of the disease. These results confirm that our genetic approach is likely to identify bona fide modulators of SMN activity, especially regarding its role at the neuromuscular junction, and as a consequence, may identify putative SMA therapeutic targets.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
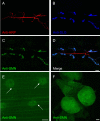
Figure 1. SMN localizes to the post-synaptic region of the Drosophila NMJ.
(A–D) SMN expression at the NMJ between muscle fibers 6 and 7. (A) Pre-synaptic anti-HRP staining (red), (B) post-synaptic anti-DLG staining (blue), (C) anti-SMN staining (green) and (D) a merge of (A–C). SMN expression co-localizes with DLG at the post-synaptic region of the NMJ. (E) SMN staining is also observed in muscle fibers and discrete foci in nuclei (arrow). (F) Though no pre-synaptic SMN staining is observed, robust levels of SMN expression are seen in the larval brain. Scale bars in (D), (E), (F) represent 10 µm, 20 µm, and 50 µm.
Figure 2. Smn mutations cause lethality.
(A) Schematic representation of the SMN protein and location of mutations corresponding to the Smn alleles used in this study. The conserved Tudor domain and YG box are indicated. Insertion sites of the transposon induced Smn f05960 and Smn f01109 alleles are denoted by triangles. Regions of the Smn transcript targeted by RNA interference (RNAi) are illustrated as lines under the SMN protein schematic. (B) Loss of Smn function elicits lethality. For individuals of given phenotypes, the percentages of surviving individuals are shown and are normalized to wild-type. Smn 73Ao and Smn f05960 homozygotes die during late 2nd/early 3rd larval and pupal stages, though some Smn f05960 escapers are detected. In contrast, 67% of the Smn f01109 homozygotes survive to adulthood. Smn f01109/Smn 73Ao and Smn f05960/Smn 73Ao trans-heterozygous combinations are also viable. In addition, a small deficiency uncovering the entire Smn transcript was generated (Df(3L)Smn X7). We crossed all three Smn alleles to Df(3L)Smn X7 and found that both Smn 73Ao/Df(3L)Smn X7 and Smn f05960/Df(3L)Smn X7 heterozygotes die between the 2nd and 3rd instar larval stages, while ∼60% of Smn f01109/Df(3L)Smn X7 are viable. Therefore, using lethality as a criterion, all three alleles behave as loss-of-function mutations with Smn f01109 displaying the weakest phenotype of the three. No obvious maternal or paternal effect is observed for the different alleles. m: maternal contribution, p: paternal contribution. WT is wild-type (Canton-S). At least 100 individuals were examined for each genotype.
Figure 3. Lethality strongly associates with loss of Smn function in muscle.
Survival rates of animals expressing the N4, C24 and FL26B transgenic _UAS_-_Smn_-RNAi constructs under the control of the actinGAL4 (A), how24BGAL4 (B), and elavGAL4 (C) drivers were measured at the following developmental stages: embryo (day 0), 1st instar larva (day 2), 3rd instar larva (day 5), early pupa (day 7), late pupa (day 9), 2-day old adult (day 12). Each experiment was performed in triplicate. The empty pWIZ RNAi vector served as a control. The survival rates of animals were calculated and subtracted from control values. The N4, C24 and FL26B transgenic animals displayed graded viability among the drivers tested. Ubiquitous SMN knockdown (A) leads to pupal lethality. Muscle-specific SMN knockdown (B) leads to late pupal lethality only in animals harboring the stronger alleles (N4 and C24), whereas greater than 90% of FL26B individuals survive to adulthood. In contrast, reduction of SMN in neurons using N4 and C24 (C) causes only very mild lethality (7%) when compared to control animals. (D) Western blots using an anti-SMN polyclonal antibody show reduction of SMN protein in 3rd instar larvae for all three _UAS_-_Smn_-RNAi transgenic strains in combination with the ubiquitous actinGAL4 driver. The top panel shows a graded effect on SMN protein levels by the three constructs consistent with their effects on lethality. The bottom panel shows anti-α tubulin levels, which served as loading controls.
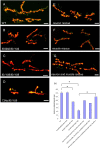
Figure 4. Drosophila Smn mutations elicit neuromuscular junction (NMJ) defects.
(A–F) The morphology of the NMJ, as judged by bouton numbers, between muscles 6 and 7 in the A2 segment was observed in different genetic backgrounds using the pre-synaptic (Synaptotagmin) and post-synaptic (Discs large) markers, shown in green and red, respectively. The following genotypes were examined: (A) wild-type (Canton-S), (B) Smn f05960/Smn f01109 (C) Smn f01109/Smn f01109, (D) Smn 73Ao/Smn f01109. Of these combinations, Smn 73Ao/Smn f01109 displayed the most robust NMJ defect. These defects are partially rescued by either (E) neuron-specific expression (elavGAL4) or (F) muscle-specific expression (how24BGAL4) of a _UAS_-_FLAG_-Smn transgene. (G) More complete rescue was achieved when this transgene was expressed using both drivers simultaneously. Bouton numbers were normalized to the ratio of the muscle area. Scale bars represent 20 µm. (H) Diagram of bouton numbers for genotypes from (A–F), normalized for muscle area. * P<0.05 was determined by the ANOVA multiple comparisons test. For each genotype at least 15 animals were examined.
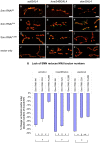
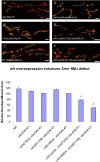
Figure 5. Muscle and neuron specific Smn RNAi knockdown causes NMJ defects.
(A–I) Reduced SMN expression in the N4, C24 and FL26B _UAS_-_Smn_-RNAi transgenic constructs elicits graded effects on NMJ morphology using the ubiquitous actinGAL4 (A, D, G) as well as the tissue-specific how24BGAL4 (muscle) (B, E, H) and elavGAL4 (neuron) (C, F, I) drivers. Vector only (pWIZ) controls are shown (J, K, L). In these images the pre- and post-synaptic tissues are labeled with antibodies against Synaptotagmin (green) and Discs large (red), respectively. (M) Bouton counts for the NMJs from the genotypes shown in (A–L) were normalized for muscle area and subtracted from vector only controls. For each genotype at least 15 animals were examined. * P<0.01 and **P<0.05 was determined by the ANOVA multiple comparisons test. Scale bars represent 15 µm.
Figure 6. Schematic representation of the Smn modifier screen.
Depicted are the crosses performed to identify enhancers and suppressors of _Smn_-associated lethality. In the first stage of the screen, designed to identify Smn enhancers, Smn 73Ao tubulinGAL4 e/TM6B virgin females were mated to males from Exelixis collection strains. In this stage, the entire Exelixis collection, which affects approximately 50% of the Drosophila genome, was tested. In the F1 generation, mutations that resulted in synthetic lethality or reduced viability in trans with the Smn 73Ao tubulinGAL4 e chromosome were defined as enhancers. In the second stage of the screen, males from F1 crosses that failed to show enhancement (_P_[Exelixis]/+; Smn 73Ao tubulinGAL4 e/TM6B) were mated to Smn 73Ao e/TM1, Mé virgin females to identify mutations that suppressed the Smn 73Ao tubulinGAL4 e/Smn 73Ao, e lethal phenotype. We performed the F2 suppressor screen with Exelixis mutations on first and second chromosomes as testing third chromosome mutations would require placing these mutations in cis with Smn. Additional assays were employed to eliminate false positives (See Materials and Methods). Seventeen enhancers and ten suppressors met these criteria. All 27 modifiers were subsequently examined for their ability to modify the Smn NMJ phenotype by GluRIIA staining (Figures S5 and S6).
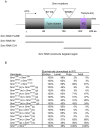
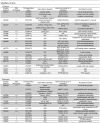
Figure 7. Modifiers of Smn phenotypes.
Listed are the insertions that enhance (top) or suppress (bottom) Smn 73Ao-dependent lethality. Due to the site of transposon insertion, unambiguous gene assignments were not possible in all instances (shaded). Strains whose designations begin with “d” or “f” contain GAL4 responsive elements (UAS), whereas strains beginning with “c” or “e” are not GAL4-inducible. Gene assignments were determined using FlyBase (
). Human homologs were determined using NCBI BLAST, NCBI UniGene (NCBI) (
http://www.ncbi.nlm.nih.gov/sites/entrezdbunigene
) or ENSEMBL genome browser (
). Annotated functions were determined based on FlyBase, NCBI Entrez Gene and SMART (
http://smart.embl-heidelberg.de/
). Modification of the NMJ morphology between muscles 6 and 7 in the A2 segment was assayed in the elavGAL4 pWIZ[_UAS-Smn-RNAi_]C24 background in trans with all identified modifiers using the pre-synaptic (Horseradish peroxidase) and post-synaptic (GluRIIA) markers (see Materials and Methods). In the three cases that did not show significant phenotypic alteration, additional pWIZ[_UAS-Smn-RNAi_]N13 allele was also used (see text). The degrees of change observed in GluRIIA staining were categorized as follows: +++, strong; ++, moderate; +, weak; N.E., No Effects.
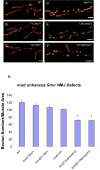
Figure 8. wit overexpression in neurons exacerbates _Smn_-dependent NMJ defects.
A gain-of-function mutation of wishful thinking (wit), wit d02492, was identified as an enhancer in our screen. To further investigate the interaction between wit and Smn at the NMJ, we used the neuron-specific driver, elavGAL4 to express WIT in neurons. (A–F) The morphology of the NMJ, as judged by bouton numbers, between muscles 6 and 7 in the A2 segment was observed in different genetic backgrounds using the pre-synaptic (Synaptotagmin) and post-synaptic (Discs large) markers, shown in green and red, respectively. The following genotypes were examined: (A) elavGAL4/+, (B) elavGAL4, Smn 73Ao/+, (C) elavGAL4, Smn f01109/+, (D) elavGAL4/UAS-wit2A, (E) elavGAL4, Smn 73Ao/UAS-wit2A, (F) elavGAL4, Smn f01109/UAS-wit2A, (G) Bouton counts for genotypes from (A–F and wild-type). Consistent with previous reports, neural induced expression of the UAS-wit2A transgene had no obvious effect on NMJ bouton number. A synergistic effect was observed upon the addition of a single Smn allele (Smn 73Ao or Smn f01109) to this background, leading to a reduction of NMJ bouton numbers. The phenotype was more severe in the Smn f01109 background. Smn f01109 showed an approximate 50% reduction in bouton numbers while Smn 73Ao reduced the bouton count by 20%. elavGAL4, Smn 73Ao/+ (B) and elavGAL4, Smn f01109/+ (C) individuals display no significant reduction in NMJ bouton numbers compared to wild-type (G). Bouton counts were determined as above. Error bars are s.e.m.; * P<0.02 was determined by the ANOVA multiple comparisons test to wild-type and all controls. n was 15–20 animals for each genotype. Bouton numbers for each genotype were normalized to the ratio of muscle areas. Scale bars represent 20 µm.
Figure 9. Loss of mad function enhances Smn NMJ defects.
(A–F) The morphology of the NMJ, as judged by bouton numbers, between muscles 6 and 7 in the A2 segment was observed in different genetic backgrounds using the pre-synaptic (Synaptotagmin) and post-synaptic (Discs large) markers, shown in green and red, respectively. The following genotypes were examined: (A) wild-type, (B) Smn 73Ao/+, (C) Smn f01109/+, (D) mad 12/+, (E) Smn 73Ao/mad 12 and (F) Smn f01109/mad 12. (G) Bouton counts for genotypes in (A–F). Introduction of mad 12 into either a Smn 73Ao/+ or a Smn f01109/+ background dominantly reduces the _Smn_-dependent NMJ bouton count. Error bars are s.e.m.; *P<0.02 was determined by the ANOVA multiple comparisons test to wild-type and all controls. n was 15–20 animals for each genotype. Bouton numbers for each genotype were normalized to the ratio of muscle areas. Scale bars represent 20 µm.
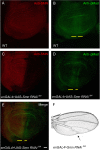
Figure 10. Smn knockdown reduces pMAD signals.
(A–B) Wild-type wing discs from 3rd instar larvae were stained with antibodies against SMN (red) (A) and phosphorylated MAD (pMAD) (green) (B). (C–D) 3rd instar wing discs of engrailedGAL4, pWIZ[_UAS-Smn-RNAi_]N4 animals are stained with antibodies against SMN (red) (C) and pMAD (green) (D). (E) Merge of (C) and (D). pMAD staining is reduced in the posterior region of the wing disc where SMN expression is decreased (yellow line). (F) A wing from an engrailedGAL4, pWIZ[_UAS-Smn-RNAi_]N4 transgenic adult exhibits defects in the posterior crossvein regions and the distal portions of wing veins L4 and L5 (arrow). Scale bars represent 40 µm.
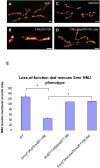
Figure 11. A dad null allele rescues Smn NMJ defects.
(A–D) The morphology of the NMJ, as judged by bouton numbers, between muscles 6 and 7 in the A2 segment was observed in different genetic backgrounds using the pre-synaptic (Synaptotagmin) and post-synaptic (Discs large) markers, shown in green and red, respectively. The following genotypes were examined: (A) wild-type (B), Smn 73Ao/Smn f01109, (C) dad 271-68 homozygotes and (D) Smn 73Ao dad 271-68/Smn f01109 dad 271-68. (E) Bouton counts for genotypes in (A–D). Smn 73Ao/Smn f01109 individuals display strongly reduced NMJ bouton numbers while dad 271-68 homozygotes have a greater than two-fold of bouton numbers relative to the Smn 73Ao/Smn f01109 animals. The Smn 73Ao dad 271-68/Smn f01109 dad 271-68 double mutants behave like dad 271-68 homozygotes. Error bars are s.e.m.; n is 15–20 animals for wild-type and Smn 73Ao/Smn f01109. n is 30 for dad 271-68/dad 271-68 and Smn 73Ao, dad 271-68/Smn f01109, dad 271-68. *Π<0.002 by the ANOVA multiple comparisons test. Bouton numbers for each genotype were normalized to the ratio of muscle area. Scale bars represent 15 µm.
Similar articles
- A critical smn threshold in mice dictates onset of an intermediate spinal muscular atrophy phenotype associated with a distinct neuromuscular junction pathology.
Bowerman M, Murray LM, Beauvais A, Pinheiro B, Kothary R. Bowerman M, et al. Neuromuscul Disord. 2012 Mar;22(3):263-76. doi: 10.1016/j.nmd.2011.09.007. Epub 2011 Nov 8. Neuromuscul Disord. 2012. PMID: 22071333 - Motor neuronal repletion of the NMJ organizer, Agrin, modulates the severity of the spinal muscular atrophy disease phenotype in model mice.
Kim JK, Caine C, Awano T, Herbst R, Monani UR. Kim JK, et al. Hum Mol Genet. 2017 Jul 1;26(13):2377-2385. doi: 10.1093/hmg/ddx124. Hum Mol Genet. 2017. PMID: 28379354 Free PMC article. - Survival motor neuron protein in motor neurons determines synaptic integrity in spinal muscular atrophy.
Martinez TL, Kong L, Wang X, Osborne MA, Crowder ME, Van Meerbeke JP, Xu X, Davis C, Wooley J, Goldhamer DJ, Lutz CM, Rich MM, Sumner CJ. Martinez TL, et al. J Neurosci. 2012 Jun 20;32(25):8703-15. doi: 10.1523/JNEUROSCI.0204-12.2012. J Neurosci. 2012. PMID: 22723710 Free PMC article. - At the "junction" of spinal muscular atrophy pathogenesis: the role of neuromuscular junction dysfunction in SMA disease progression.
Goulet BB, Kothary R, Parks RJ. Goulet BB, et al. Curr Mol Med. 2013 Aug;13(7):1160-74. doi: 10.2174/15665240113139990044. Curr Mol Med. 2013. PMID: 23514457 Review. - New and Developing Therapies in Spinal Muscular Atrophy: From Genotype to Phenotype to Treatment and Where Do We Stand?
Chen TH. Chen TH. Int J Mol Sci. 2020 May 7;21(9):3297. doi: 10.3390/ijms21093297. Int J Mol Sci. 2020. PMID: 32392694 Free PMC article. Review.
Cited by
- SMA-MAP: a plasma protein panel for spinal muscular atrophy.
Kobayashi DT, Shi J, Stephen L, Ballard KL, Dewey R, Mapes J, Chung B, McCarthy K, Swoboda KJ, Crawford TO, Li R, Plasterer T, Joyce C; Biomarkers for Spinal Muscular Atrophy Study Group; Chung WK, Kaufmann P, Darras BT, Finkel RS, Sproule DM, Martens WB, McDermott MP, De Vivo DC; Pediatric Neuromuscular Clinical Research Network; Walker MG, Chen KS. Kobayashi DT, et al. PLoS One. 2013;8(4):e60113. doi: 10.1371/journal.pone.0060113. Epub 2013 Apr 2. PLoS One. 2013. PMID: 23565191 Free PMC article. - Spinal muscular atrophy: new and emerging insights from model mice.
Park GH, Kariya S, Monani UR. Park GH, et al. Curr Neurol Neurosci Rep. 2010 Mar;10(2):108-17. doi: 10.1007/s11910-010-0095-5. Curr Neurol Neurosci Rep. 2010. PMID: 20425235 Free PMC article. Review. - Analysis of asymptomatic Drosophila models for ALS and SMA reveals convergent impact on functional protein complexes linked to neuro-muscular degeneration.
Garcia-Vaquero ML, Heim M, Flix B, Pereira M, Palin L, Marques TM, Pinto FR, de Las Rivas J, Voigt A, Besse F, Gama-Carvalho M. Garcia-Vaquero ML, et al. BMC Genomics. 2023 Sep 27;24(1):576. doi: 10.1186/s12864-023-09562-4. BMC Genomics. 2023. PMID: 37759179 Free PMC article. - Amyotrophic Lateral Sclerosis Modifiers in Drosophila Reveal the Phospholipase D Pathway as a Potential Therapeutic Target.
Kankel MW, Sen A, Lu L, Theodorou M, Dimlich DN, McCampbell A, Henderson CE, Shneider NA, Artavanis-Tsakonas S. Kankel MW, et al. Genetics. 2020 Jul;215(3):747-766. doi: 10.1534/genetics.119.302985. Epub 2020 Apr 28. Genetics. 2020. PMID: 32345615 Free PMC article. - Generation and Characterization of a genetic zebrafish model of SMA carrying the human SMN2 gene.
Hao le T, Burghes AH, Beattie CE. Hao le T, et al. Mol Neurodegener. 2011 Mar 28;6(1):24. doi: 10.1186/1750-1326-6-24. Mol Neurodegener. 2011. PMID: 21443782 Free PMC article.
References
- Monani UR. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48:885–896. - PubMed
- Frugier T, Nicole S, Cifuentes-Diaz C, Melki J. The molecular bases of spinal muscular atrophy. Curr Opin Genet Dev. 2002;12:294–298. - PubMed
- Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum Mutat. 2000;15:228–237. - PubMed
- Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis. 1996;3:97–110. - PubMed
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous