The evolving transcriptome of head and neck squamous cell carcinoma: a systematic review - PubMed (original) (raw)
Meta-Analysis
The evolving transcriptome of head and neck squamous cell carcinoma: a systematic review
Yau-Hua Yu et al. PLoS One. 2008.
Abstract
Background: Numerous studies were performed to illuminate mechanisms of tumorigenesis and metastases from gene expression profiles of Head and Neck Squamous Cell Carcinoma (HNSCC). The objective of this review is to conduct a network-based meta-analysis to identify the underlying biological signatures of the HNSCC transcriptome.
Methods and findings: We included 63 HNSCC transcriptomic studies into three specific categories of comparisons: Pre, premalignant lesions v.s. normal; TvN, primary tumors v.s. normal; and Meta, metastatic or invasive v.s. primary tumors. Reported genes extracted from the literature were systematically analyzed. Participation of differential gene activities across three progressive stages deciphered the evolving nature of HNSCC. In total, 1442 genes were verified, i.e. reported at least twice, with ECM1, EMP1, CXCL10 and POSTN shown to be highly reported across all three stages. Knowledge-based networks of the HNSCC transcriptome were constructed, demonstrating integrin signaling and antigen presentation pathways as highly enriched. Notably, functional estimates derived from topological characteristics of integrin signaling networks identified such important genes as ITGA3 and ITGA5, which were supported by findings of invasiveness in vitro. Moreover, we computed genome-wide probabilities of reporting differential gene activities for the Pre, TvN, and Meta stages, respectively. Results highlighted chromosomal regions of 6p21, 19p13 and 19q13, where genomic alterations were shown to be correlated with the nodal status of HNSCC.
Conclusions: By means of a systems-biology approach via network-based meta-analyses, we provided a deeper insight into the evolving nature of the HNSCC transcriptome. Enriched canonical signaling pathways, hot-spots of transcriptional profiles across the genome, as well as topologically significant genes derived from network analyses were highlighted for each of the three progressive stages, Pre, TvN, and Meta, respectively.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
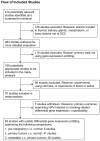
Figure 1. QUOROM flow diagram of the systematic reviews and meta-analysis.
The diagram summarized the search strategy. In order to be included, studies had to examine the HNSCC tumor samples by means of microarray-based gene expression profiling.
Figure 2. Ranking of the enriched canonical pathways.
Eight canonical pathways were found in consensus in IPA with the full gene lists (all) or the verified gene lists (fq2) as inputs. Embedded table showed rankings of the canonical pathways in different analyses. Antigen presentation, calcium signaling, and integrin signaling pathways were highlighted as highly enriched across three progressive stages. For each stage of the HNSCC transcriptome, figure panels demonstrated the enriched canonical pathways ranked by the −log (p-values) (right y-axis), that is, the orange line with square data points. Colored bars were indicating the percentage (left y-axis) of the up- or down-regulated genes within each canonical pathway. The numbers on top of the colored bars were the number of total genes in the canonical pathways.
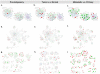
Figure 3. Enriched antigen presentation and integrin signaling pathways in each stage of comparison of the HNSCC transcriptome.
Genes were represented by nodes and functional associations by edges. Node coloring was scaled to the bounded fold changes–red: up-regulated; green: down-regulated. Node size was proportional to the number of papers reporting this gene in the HNSCC transriptome. Edges were colored according to the stage of the HNSCC transcriptome–red: Meta; blue: TvN; and green: Pre. a–c. Merged networks of the antigen presentation pathways with nodes colored according to the bounded fold changes in Pre, TvN, and Meta, respectively. d–f. Merged networks of the integrin signaling networks with nodes colored according to the bounded fold changes in Pre, TvN, and Meta, respectively. g–i. Sub-networks of the integrin signaling pathway in the Pre, TvN and Meta stages of the HNSCC transcriptome. (For details, please see Figure S1, S2, S3, S4 and S5).
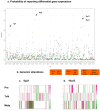
Figure 4. Genome-wide probabilities of the evolving HNSCC transcriptome.
a. We computed the accumulative probabilities of reporting differentially expressed genes within each cytoband along the chromosomal coordinates. Hot-spots of differential gene activities, 1p21, 6p21, 19p13 and 19q13, were highlighted. The standardized accumulative probability for each cytoband was plotted across the genome. The color schemes represented three progressive stages of the HNSCC transcriptome–red circles: Meta; blue squares: TvN; and green triangles: Pre. b. Co-localization between hot-spots of differential gene activities and previously implicated regions of genomic alterations by Weber et al. . c–d. Differential gene expression profiles–bounded fold changes - of 167 genes in 6p21 and 189 genes in 19q13 were plotted along the chromosomal coordinates for the Pre, TvN and Meta stages, respectively.
Similar articles
- Deep sequencing-based microRNA expression signatures in head and neck squamous cell carcinoma: dual strands of pre-miR-150 as antitumor miRNAs.
Koshizuka K, Nohata N, Hanazawa T, Kikkawa N, Arai T, Okato A, Fukumoto I, Katada K, Okamoto Y, Seki N. Koshizuka K, et al. Oncotarget. 2017 May 2;8(18):30288-30304. doi: 10.18632/oncotarget.16327. Oncotarget. 2017. PMID: 28415821 Free PMC article. - Differential gene expression signature between primary and metastatic head and neck squamous cell carcinoma.
Liu CJ, Liu TY, Kuo LT, Cheng HW, Chu TH, Chang KW, Lin SC. Liu CJ, et al. J Pathol. 2008 Mar;214(4):489-97. doi: 10.1002/path.2306. J Pathol. 2008. PMID: 18213732 - Transcriptome analysis reveals the link between lncRNA-mRNA co-expression network and tumor immune microenvironment and overall survival in head and neck squamous cell carcinoma.
Zhong Z, Hong M, Chen X, Xi Y, Xu Y, Kong D, Deng J, Li Y, Hu R, Sun C, Liang J. Zhong Z, et al. BMC Med Genomics. 2020 Mar 30;13(1):57. doi: 10.1186/s12920-020-0707-0. BMC Med Genomics. 2020. PMID: 32228580 Free PMC article. - The Notch signaling pathway in head and neck squamous cell carcinoma: A meta-analysis.
Zhao YY, Yu GT, Xiao T, Hu J. Zhao YY, et al. Adv Clin Exp Med. 2017 Aug;26(5):881-887. doi: 10.17219/acem/64000. Adv Clin Exp Med. 2017. PMID: 29068587 Review. - Integrins in head and neck squamous cell carcinoma (HNSCC): a review of the current literature.
Georgolios AK, Batistatou A, Charalabopoulos K. Georgolios AK, et al. Cell Commun Adhes. 2005 Jan-Apr;12(1-2):1-8. doi: 10.1080/15419060500383093. Cell Commun Adhes. 2005. PMID: 16371342 Review.
Cited by
- Extra-Capsular Spread of Lymph Node Metastasis in Oral, Oropharyngeal and Hypopharyngeal Cancer: A Comparative Subsite Analysis.
Kang YJ, Park G, Park SY, Kim T, Kim E, Heo Y, Lee C, Jeong HS. Kang YJ, et al. Cancers (Basel). 2024 Feb 3;16(3):659. doi: 10.3390/cancers16030659. Cancers (Basel). 2024. PMID: 38339410 Free PMC article. - Decoding tumor heterogeneity in uveal melanoma: basement membrane genes as novel biomarkers and therapeutic targets revealed by multi-omics approaches for cancer immunotherapy.
Li Y, Cai H, Yang J, Xie X, Pei S, Wu Y, Zhang J, Song G, Zhang J, Zhang Q, Chi H, Yang G. Li Y, et al. Front Pharmacol. 2023 Sep 26;14:1264345. doi: 10.3389/fphar.2023.1264345. eCollection 2023. Front Pharmacol. 2023. PMID: 37822877 Free PMC article. - Expression of EMP 1, 2, and 3 in Adrenal Cortical Neoplasm and Pheochromocytoma.
Cha YJ, Koo JS. Cha YJ, et al. Int J Mol Sci. 2023 Aug 21;24(16):13016. doi: 10.3390/ijms241613016. Int J Mol Sci. 2023. PMID: 37629198 Free PMC article. - Prognostic-related genes for pancreatic cancer typing and immunotherapy response prediction based on single-cell sequencing data and bulk sequencing data.
Wang X, Jiang S, Zhou X, Wang X, Li L, Tang J. Wang X, et al. Oncol Res. 2023 Jul 21;31(5):697-714. doi: 10.32604/or.2023.029458. eCollection 2023. Oncol Res. 2023. PMID: 37547756 Free PMC article. - Transcriptomic analysis predicts the risk of progression of premalignant lesions in human tongue.
Zhang T, Kutler D, Scognamiglio T, Gudas LJ, Tang XH. Zhang T, et al. Discov Oncol. 2023 Feb 23;14(1):24. doi: 10.1007/s12672-023-00629-y. Discov Oncol. 2023. PMID: 36820942 Free PMC article.
References
- Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392–1400. - PubMed
- Weber F, Xu Y, Zhang L, Patocs A, Shen L, et al. Microenvironmental genomic alterations and clinicopathological behavior in head and neck squamous cell carcinoma. Jama. 2007;297:187–195. - PubMed
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. - PubMed
- Seiwert TY, Salama JK, Vokes EE. The chemoradiation paradigm in head and neck cancer. Nat Clin Pract Oncol. 2007;4:156–171. - PubMed
- Choi P, Chen C. Genetic expression profiles and biologic pathway alterations in head and neck squamous cell carcinoma. Cancer. 2005;104:1113–1128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous