Loss of Hus1 sensitizes cells to etoposide-induced apoptosis by regulating BH3-only proteins - PubMed (original) (raw)
Loss of Hus1 sensitizes cells to etoposide-induced apoptosis by regulating BH3-only proteins
C L Meyerkord et al. Oncogene. 2008.
Abstract
The Rad9-Rad1-Hus1 (9-1-1) cell cycle checkpoint complex plays a key role in the DNA damage response. Cells with a defective 9-1-1 complex have been shown to be sensitive to apoptosis induced by certain types of genotoxic stress. However, the mechanism linking the loss of a functional 9-1-1 complex to the cell death machinery has yet to be determined. Here, we report that etoposide treatment dramatically upregulates the BH3-only proteins, Bim and Puma, in Hus1-deficient cells. Inhibition of either Bim or Puma expression in Hus1-knockout cells confers significant resistance to etoposide-induced apoptosis, whereas knockdown of both proteins results in further resistance, suggesting that Bim and Puma cooperate in sensitizing Hus1-deficient cells to etoposide treatment. Moreover, we found that Rad9 collaborates with Bim and Puma to sensitize Hus1-deficient cells to etoposide-induced apoptosis. In response to DNA damage, Rad9 localizes to chromatin in Hus1-wild-type cells, whereas in Hus1-deficient cells, it is predominantly located in the cytoplasm where it binds to Bcl-2. Taken together, these results suggest that loss of Hus1 sensitizes cells to etoposide-induced apoptosis not only by inducing Bim and Puma expressions but also by releasing Rad9 into the cytosol to augment mitochondrial apoptosis.
Figures
Figure 1
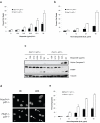
Loss of Hus1 sensitizes cells to apoptosis induced by etoposide treatment. (a) Hus1+/+_p21_-/- and _Hus1_-/- _p21_-/- MEFs were treated with increasing doses of etoposide for 24 h and viability was determined by trypan blue exclusion assay (mean ± s.d.; n=3). (b) Hus1+/+_p21_-/- and _Hus1_-/-_p21_-/- MEFs were treated with 6.25 μg/ml etoposide for varying time points and subjected to trypan blue exclusion assay (mean ± s.d.; n=3). (c) Cells were treated as in (a). Total cell lysate was normalized for protein content and subjected to SDS-PAGE/immunoblot analysis. (d) Hus1+/+_p21_-/- and _Hus1_-/-_p21_-/- MEFs were treated with 6.25 μg/ml etoposide for 0 or 48 h. Apoptosis was determined by examination of nuclear morphology. Arrows indicate apoptotic nuclei. (e) Hus1+/+_p21_-/- and _Hus1_-/-_p21_-/- MEFs were treated with 6.25 μg/ml etoposide for the times indicated. The percent of apoptotic cells was quantified based on nuclear morphology (mean ± s.d.; n=3).
Figure 2
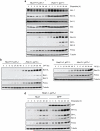
Loss of Hus1 results in upregulation of Bim and Puma expression in response to DNA damage. (a, b, c) Hus1+/+_p21_-/- and _Hus1_-/-_p21_-/- MEFs were treated with (a) 6.25 μg/ml etoposide, (b) 500 nM camptothecin (CPT) or (c) 50 μM hydroxyurea (HU) for the indicated time points. Total cell lysate was prepared and analyzed by SDS-PAGE/immunoblot using the indicated antibodies. (d) _Hus1_-/-_p21_-/- MEFs stably expressing Hus1 or GFP were treated with 6.25 μg/ml etoposide for varying time points. The expression of Bim and Puma was examined by SDS-PAGE/immunoblot analysis.
Figure 3
Induction of Bim and Puma expression in response to etoposide treatment is regulated at the transcriptional level. (a) _Hus1_-/-_p21_-/- MEFs were treated with control DMSO (-), 1 μg/ml actinomycin D or 5 μg/ml cycloheximide alone or in combination with 3.125 μg/ml etoposide for 24 h. Total cell lysate was prepared and the expression of Bim and Puma was analyzed by SDS-PAGE/immunoblot. (b, c) Hus1+/+_p21_-/- and _Hus1_-/-_p21_-/- MEFs were treated with 6.25 μg/ml etoposide for 0, 3, 6, 12, 24, 36 or 48 h. Semi-quantitative RT-PCR was used to examine the mRNA levels of (b) Bim and (c) Puma. (d, e) Hus1+/+_p21_-/- and _Hus1_-/-_p21_-/- MEFs were treated with 25 μg/ml etoposide for 0, 1, 2, 3 or 4 h. Semi-quantitative RT-PCR was used to examine the mRNA levels of (d) Bim and (e) Puma.
Figure 4
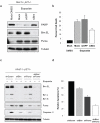
Suppression of Bim and Puma expression confers resistance to etoposide-induced apoptosis in _Hus1_-deficient cells. (a, b) _Hus1_-/-_p21_-/- MEFs were mock transfected or transiently transfected with siRNA targeting GFP or Bim. Thirty-six hours after transfection, the cells were treated with control DMSO or 6.25 μg/ml etoposide for 30 h. (a) Whole cell lysate was subjected to SDS-PAGE/immunoblot analysis with antibodies to PARP (full length PARP is shown), Bim, Puma and Tubulin. (b) Viability was determined by trypan blue exclusion assay (mean ± s.d.; n=2). The DMSO-treated mock transfected cells were used to show the basal levels of Bim expression and cell death. (c, d) _Hus1_-/-_p21_-/- MEFs were infected with lentivirus expressing shRNA targeting Bim, Puma, Bim and Puma, or a control scrambled shRNA (shScram). After selection on Puromycin, the cells were treated with 12.5 μg/ml etoposide or control DMSO for 16 h. (c) Knockdown of Bim and Puma was confirmed by SDS-PAGE/immunoblot analysis. (d) Induction of apoptosis was measured by caspase-3 activity assay. The caspase-3 activity of control DMSO treated cells was subtracted from the amount of caspase-3 activity observed in the etoposide treated cells. The data are represented as percent relative apoptosis as normalized to the control infected cells (mean ± s.d.; n=3).
Figure 5
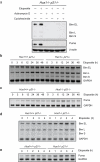
Rad9 is predominantly detected in the cytoplasm of _Hus1_-deficient cells where it binds to Bcl-2 in response to DNA damage. (a) Hus1+/+_p21_-/- and _Hus1_-/-_p21_-/- MEFs were treated with 12.5 μg/ml etoposide for 0, 2 or 8 h and subjected to subcellular fractionation. The resulting chromatin bound and soluble fractions were analyzed by SDS-PAGE/immunoblot using antibodies specific for Rad9 and RPA as a control. (b) Hus1+/+_p21_-/- and _Hus1_-/-_p21_-/- MEFs were treated with 12.5 μg/ml etoposide or control DMSO for 12 h and subjected to subcellular fractionation. The resulting cytosolic (Cyt) and nuclear (Nuc) fractions, along with whole cell lysate (WCL), were analyzed by SDS-PAGE/immunoblot. (c, d) Hus1+/+_p21_-/- and _Hus1_-/-_p21_-/- MEFs were treated with 12.5 μg/ml etoposide or control DMSO for 12 h. (c) The cytosolic fractions of Hus1+/+_p21_-/- and _Hus1_-/-_p21_-/- MEFs were subjected to immunoprecipitation in the absence or presence of anti-Bcl-2 monoclonal antibody. The resulting immune complexes were analyzed by SDS-PAGE/immunoblot. (d) Whole cell lysate (WCL) was subjected to immunoprecipitation with anti-Bcl-2 or control anti-Flag monoclonal antibodies. The resulting immunocomplexes and WCL were analyzed by SDS-PAGE/immunoblot. The amount of Rad9 in the immunocomplexes was quantified and normalized to cytosolic (c) or total (d) Rad9. The levels of Bcl-2 bound Rad9 are listed relative to those of untreated Hus1-/-p21-/- cells, which were set as 1.0.
Figure 6
Rad9 collaborates with Bim and Puma to sensitize _Hus1_-deficient cells to etoposide-induced apoptosis. (a, b) _Hus1_-/-_p21_-/- MEFs stably expressing shBim and shPuma were infected with lentivirus expressing shRNA targeting Rad9. The cells were treated with 12.5 μg/ml etoposide or control DMSO for 16 h. (a) Knockdown of Rad9, as well as Bim and Puma, was confirmed by SDS-PAGE/immunoblot analysis. (b) Induction of apoptosis was measured using a caspase-3 activity assay. (c) _Hus1_-/-_p21_-/- MEFs stably expressing control shRNA, shBim and shPuma, or shBim and shPuma plus shRad9 were treated with 12.5 μg/ml etoposide for 0, 24 or 48 h. Cell death was determined by trypan blue exclusion assay (mean ± s.d.; n=3).
Similar articles
- The BH3-only protein Puma plays an essential role in cytokine deprivation induced apoptosis of mast cells.
Ekoff M, Kaufmann T, Engström M, Motoyama N, Villunger A, Jönsson JI, Strasser A, Nilsson G. Ekoff M, et al. Blood. 2007 Nov 1;110(9):3209-17. doi: 10.1182/blood-2007-02-073957. Epub 2007 Jul 18. Blood. 2007. PMID: 17634411 Free PMC article. - The effect of ionizing radiation on mRNA levels of the DNA damage response genes rad9, rad1 and hus1 in various mouse tissues.
Zhang Z, Cai Z, Li K, Fang Y, An L, Hu Z, Wang S, Hang H. Zhang Z, et al. Radiat Res. 2015 Jan;183(1):94-104. doi: 10.1667/RR13781.1. Epub 2015 Jan 7. Radiat Res. 2015. PMID: 25564717 - BH3-only proteins Puma and Bim are rate-limiting for gamma-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo.
Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L, Adams JM, Strasser A, Villunger A. Erlacher M, et al. Blood. 2005 Dec 15;106(13):4131-8. doi: 10.1182/blood-2005-04-1595. Epub 2005 Aug 23. Blood. 2005. PMID: 16118324 Free PMC article. - Malignant mesothelioma cells are rapidly sensitized to TRAIL-induced apoptosis by low-dose anisomycin via Bim.
Abayasiriwardana KS, Barbone D, Kim KU, Vivo C, Lee KK, Dansen TB, Hunt AE, Evan GI, Broaddus VC. Abayasiriwardana KS, et al. Mol Cancer Ther. 2007 Oct;6(10):2766-76. doi: 10.1158/1535-7163.MCT-07-0278. Mol Cancer Ther. 2007. PMID: 17938269 - Contributions of Rad9 to tumorigenesis.
Broustas CG, Lieberman HB. Broustas CG, et al. J Cell Biochem. 2012 Mar;113(3):742-51. doi: 10.1002/jcb.23424. J Cell Biochem. 2012. PMID: 22034047 Free PMC article. Review.
Cited by
- The carboxyl-terminal sequence of PUMA binds to both anti-apoptotic proteins and membranes.
Pemberton JM, Nguyen D, Osterlund EJ, Schormann W, Pogmore JP, Hirmiz N, Leber B, Andrews DW. Pemberton JM, et al. Elife. 2023 Apr 20;12:e88329. doi: 10.7554/eLife.88329. Elife. 2023. PMID: 37078707 Free PMC article. - Prostate cancer: unmet clinical needs and RAD9 as a candidate biomarker for patient management.
Lieberman HB, Rai AJ, Friedman RA, Hopkins KM, Broustas CG. Lieberman HB, et al. Transl Cancer Res. 2018 Jul;7(Suppl 6):S651-S661. doi: 10.21037/tcr.2018.01.21. Epub 2018 Jan 14. Transl Cancer Res. 2018. PMID: 30079300 Free PMC article. - ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands.
Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. Welboren WJ, et al. EMBO J. 2009 May 20;28(10):1418-28. doi: 10.1038/emboj.2009.88. Epub 2009 Apr 4. EMBO J. 2009. PMID: 19339991 Free PMC article. - Mutation to Bax beyond the BH3 domain disrupts interactions with pro-survival proteins and promotes apoptosis.
Czabotar PE, Lee EF, Thompson GV, Wardak AZ, Fairlie WD, Colman PM. Czabotar PE, et al. J Biol Chem. 2011 Mar 4;286(9):7123-31. doi: 10.1074/jbc.M110.161281. Epub 2011 Jan 3. J Biol Chem. 2011. PMID: 21199865 Free PMC article. - HUS1 as a Potential Therapeutic Target in Urothelial Cancer.
Lindner AK, Furlan T, Orme JJ, Tulchiner G, Staudacher N, D'Andrea D, Culig Z, Pichler R. Lindner AK, et al. J Clin Med. 2022 Apr 15;11(8):2208. doi: 10.3390/jcm11082208. J Clin Med. 2022. PMID: 35456300 Free PMC article.
References
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–96. - PubMed
- Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. - PubMed
- Bao S, Lu T, Wang X, Zheng H, Wang LE, Wei Q, et al. Disruption of the Rad9/Rad1/Hus1 (9-1-1) complex leads to checkpoint signaling and replication defects. Oncogene. 2004;23:5586–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA90315/CA/NCI NIH HHS/United States
- R01 CA090315-05/CA/NCI NIH HHS/United States
- CA108773/CA/NCI NIH HHS/United States
- R01 CA108773/CA/NCI NIH HHS/United States
- R01 CA090315/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials