CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity - PubMed (original) (raw)
. 2008 Sep 25;455(7212):547-51.
doi: 10.1038/nature07179. Epub 2008 Sep 14.
Adam J Bass, So Young Kim, Ian F Dunn, Serena J Silver, Isil Guney, Ellen Freed, Azra H Ligon, Natalie Vena, Shuji Ogino, Milan G Chheda, Pablo Tamayo, Stephen Finn, Yashaswi Shrestha, Jesse S Boehm, Supriya Jain, Emeric Bojarski, Craig Mermel, Jordi Barretina, Jennifer A Chan, Jose Baselga, Josep Tabernero, David E Root, Charles S Fuchs, Massimo Loda, Ramesh A Shivdasani, Matthew Meyerson, William C Hahn
Affiliations
- PMID: 18794900
- PMCID: PMC2587138
- DOI: 10.1038/nature07179
CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity
Ron Firestein et al. Nature. 2008.
Abstract
Aberrant activation of the canonical WNT/beta-catenin pathway occurs in almost all colorectal cancers and contributes to their growth, invasion and survival. Although dysregulated beta-catenin activity drives colon tumorigenesis, further genetic perturbations are required to elaborate full malignant transformation. To identify genes that both modulate beta-catenin activity and are essential for colon cancer cell proliferation, we conducted two loss-of-function screens in human colon cancer cells and compared genes identified in these screens with an analysis of copy number alterations in colon cancer specimens. One of these genes, CDK8, which encodes a member of the mediator complex, is located at 13q12.13, a region of recurrent copy number gain in a substantial fraction of colon cancers. Here we show that the suppression of CDK8 expression inhibits proliferation in colon cancer cells characterized by high levels of CDK8 and beta-catenin hyperactivity. CDK8 kinase activity was necessary for beta-catenin-driven transformation and for expression of several beta-catenin transcriptional targets. Together these observations suggest that therapeutic interventions targeting CDK8 may confer a clinical benefit in beta-catenin-driven malignancies.
Figures
Figure 1. RNAi screens to identify genes essential for colon cancer cell proliferation and β-catenin activity
(a) Schematic of the DLD1Rep cell line showing the engineered 8X TOPFLASH and 8X FOPFLASH elements and relative TOP/FOP activity in the DLD1Rep cell line. (b) Distribution curve showing Z-scores representing β-catenin activity for all shRNAs tested in the DLD1Rep screen. shRNAs that reduced FOPFLASH levels to near background or activated FOPFLASH more than 2 standard deviations (SD) above mean were excluded (FOP ≤800 and FOP ≥2600 luciferase units). 2 of 5 _CDK8_-specific shRNAs were excluded on this basis. shRNA that induced Z-scores > 4 are not shown. Dashed line indicates Z score cutoff for shRNAs scored as hits. (c) Distribution curve showing Z-scores representing cell proliferation for shRNAs tested in HCT116 cells. This screen contained shRNAs targeting 1004 genes, and there was 92% overlap between the screens in (b) and (c). Blue and black dashed lines indicates Z score cutoff for shRNAs scored as hits. (d) Venn Diagram representation of 9 genes that reduced both β-catenin activity and colon cancer cell proliferation.
Figure 2. Amplification and overexpression of CDK8 defines a subset of colon cancers
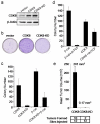
(a) Significance (y-axis) of recurrent amplifications at loci across the 22 autosomes (x-axis) identified by GISTIC analysis. Chromosomal location of RNAi hits is indicated above plot. (b) Heat map showing clustering of SNP array data, based on chromosome 13q12 copy number in 123 colon cancer specimens. Red indicates allelic gain, blue denotes loss. (c) Pie chart depicts the percentage of colon tumors exhibiting CDK8 CN gain, chromosome 13 polysomy or disomy (n=50) by FISH. Cutoff criteria for FISH are shown in Supplementary Table 4. (d) GSEA comparative analysis of suppressing resident genes in the minimal 13q12 region. Blue lines represent differential scores of cell proliferation effects for each validated shRNA that suppressed target gene expression by greater than 70%. The leftmost column demonstrates the total pool of validated hairpins for all genes on 13q12. Red lines represent a normalized enrichment score for each gene that takes into account the cell proliferation effects of all shRNAs and scores the specificity of the effects in cell lines that harbor or lack 13q gain. (e-i) Immunoblot analysis of CDK8 expression in 12 colon cancer cell lines. β-actin serves as a loading control (e). Effect of CDK8 suppression on proliferation of colon cancer cells that harbor chromosome 13 gain (f), harbor chromosome 13 loss (g), are disomic with higher CDK8 protein expression (h) and are disomic with lower CDK8 protein expression (i). Bar graph depicts cell proliferation normalized to the shGFP control for triplicate determinations. Error bars represent mean ± SD for a representative experiment performed in triplicate.
Figure 3. CDK8 and transformation
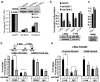
(a) Immunoblot analysis showing CDK8 and CDK8 kinase-dead (CDK8-KD) expression in NIH-3T3 cells. (b) Focus formation assay of NIH-3T3 cells expressing CDK8 or CDK8-KD. (c) CDK8 kinase activity drives anchorage independent (AI) growth and is necessary for β-catenin mediated AI growth. AI colony growth in NIH-3T3 cells infected with the indicated retroviral vectors. (d) β-catenin suppression only partial blocks CDK8 induced AI growth. Dominant negative TCF (dnTCF) was introduced in the presence of β-catenin or CDK8. AI colony growth was performed as indicated in (c). (e) CDK8 kinase activity drives tumor formation. Mean tumor volume from subcutaneous tumors formed NIH-3T3 cells expressing CDK8 or CDK8-KD constructs in immunodeficient mice. The difference in tumor formation between CDK8 and CDK8-KD was statistically significant, as assessed by unpaired t-test (p value = 0.0001). All experiments were performed in triplicate, and mean ± SD is shown.
Figure 4. CDK8 mediates transcription of β-catenin driven downstream target genes
(a) Bar graph depicts β-catenin activity as normalized TOPFLASH (TOPFLASH/FOPFLASH) after introduction of _CDK8_-specific shRNAs. An immunoblot shows CDK8 protein levels at time of assay. β-actin serves as a loading control. (b, c) Bar graphs shows mRNA abundance of endogenous β-catenin and Notch transcriptional targets after CDK8 suppression in COLO-205 and DLD1 cells. Myc, β-catenin, and Notch expression changes were assessed by immunoblotting. β-actin expression serves as a loading control. (d) CDK8 binds the MYC promoter. Schematic representation depicts the location of proximal and distal TCF binding elements (TBE, gray boxes) and MYC promoter TATA box. Dashed Lines indicate the distance from TBEs to TATA box. Hatched lines depict the PCR primers used for chromatin immunoprecipitation (ChIP). Bar graph shows CDK8 binding to the MYC TATA box in COLO-205 colon cancer cells treated with the indicated shRNAs and assayed by ChIP. (d, e) Asterisk indicates statistically significance as assessed by unpaired student t-test (p value ≤ 0.013) and p ≤ 0.14 for the control IgG for (d) and (p value ≤ 0.005) and p ≤ 0.34 for the IgG control. For (e). (e) Bar graph depicts β-catenin binding to the indicated MYC promoter elements in the presence or absence of CDK8. ChIP assays were performed as above. All experiments were conducted at least two times in triplicate. Error bars represent the mean ± SD for a representative experiment performed in triplicate. Asterisk indicates a statistically significance as assessed by unpaired student t-test
Comment in
- Cancer: Entangled pathways.
Bernards R. Bernards R. Nature. 2008 Sep 25;455(7212):479-80. doi: 10.1038/455479a. Nature. 2008. PMID: 18818647 No abstract available.
Similar articles
- CDK8 expression in 470 colorectal cancers in relation to beta-catenin activation, other molecular alterations and patient survival.
Firestein R, Shima K, Nosho K, Irahara N, Baba Y, Bojarski E, Giovannucci EL, Hahn WC, Fuchs CS, Ogino S. Firestein R, et al. Int J Cancer. 2010 Jun 15;126(12):2863-73. doi: 10.1002/ijc.24908. Int J Cancer. 2010. PMID: 19790197 Free PMC article. - Role of CDK8 and beta-catenin in colorectal adenocarcinoma.
Seo JO, Han SI, Lim SC. Seo JO, et al. Oncol Rep. 2010 Jul;24(1):285-91. Oncol Rep. 2010. PMID: 20514474 - H19 Noncoding RNA, an Independent Prognostic Factor, Regulates Essential Rb-E2F and CDK8-β-Catenin Signaling in Colorectal Cancer.
Ohtsuka M, Ling H, Ivan C, Pichler M, Matsushita D, Goblirsch M, Stiegelbauer V, Shigeyasu K, Zhang X, Chen M, Vidhu F, Bartholomeusz GA, Toiyama Y, Kusunoki M, Doki Y, Mori M, Song S, Gunther JR, Krishnan S, Slaby O, Goel A, Ajani JA, Radovich M, Calin GA. Ohtsuka M, et al. EBioMedicine. 2016 Nov;13:113-124. doi: 10.1016/j.ebiom.2016.10.026. Epub 2016 Oct 19. EBioMedicine. 2016. PMID: 27789274 Free PMC article. - Revving the Throttle on an oncogene: CDK8 takes the driver seat.
Firestein R, Hahn WC. Firestein R, et al. Cancer Res. 2009 Oct 15;69(20):7899-901. doi: 10.1158/0008-5472.CAN-09-1704. Epub 2009 Oct 6. Cancer Res. 2009. PMID: 19808961 Free PMC article. Review. - Wogonin induced G1 cell cycle arrest by regulating Wnt/β-catenin signaling pathway and inactivating CDK8 in human colorectal cancer carcinoma cells.
He L, Lu N, Dai Q, Zhao Y, Zhao L, Wang H, Li Z, You Q, Guo Q. He L, et al. Toxicology. 2013 Oct 4;312:36-47. doi: 10.1016/j.tox.2013.07.013. Epub 2013 Jul 30. Toxicology. 2013. PMID: 23907061 Review.
Cited by
- Baculovirus expression: tackling the complexity challenge.
Barford D, Takagi Y, Schultz P, Berger I. Barford D, et al. Curr Opin Struct Biol. 2013 Jun;23(3):357-64. doi: 10.1016/j.sbi.2013.03.009. Epub 2013 Apr 27. Curr Opin Struct Biol. 2013. PMID: 23628287 Free PMC article. Review. - Small molecule inhibitors of transcriptional cyclin-dependent kinases impose HIV-1 latency, presenting "block and lock" treatment strategies.
Horvath RM, Brumme ZL, Sadowski I. Horvath RM, et al. Antimicrob Agents Chemother. 2024 Mar 6;68(3):e0107223. doi: 10.1128/aac.01072-23. Epub 2024 Feb 6. Antimicrob Agents Chemother. 2024. PMID: 38319085 Free PMC article. - Cyclin-Dependent Kinases 8 and 19 Regulate Host Cell Metabolism during Dengue Virus Serotype 2 Infection.
Butler M, Chotiwan N, Brewster CD, DiLisio JE, Ackart DF, Podell BK, Basaraba RJ, Perera R, Quackenbush SL, Rovnak J. Butler M, et al. Viruses. 2020 Jun 17;12(6):654. doi: 10.3390/v12060654. Viruses. 2020. PMID: 32560467 Free PMC article. - CDX2 is an amplified lineage-survival oncogene in colorectal cancer.
Salari K, Spulak ME, Cuff J, Forster AD, Giacomini CP, Huang S, Ko ME, Lin AY, van de Rijn M, Pollack JR. Salari K, et al. Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):E3196-205. doi: 10.1073/pnas.1206004109. Epub 2012 Oct 29. Proc Natl Acad Sci U S A. 2012. PMID: 23112155 Free PMC article. - CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response.
Bancerek J, Poss ZC, Steinparzer I, Sedlyarov V, Pfaffenwimmer T, Mikulic I, Dölken L, Strobl B, Müller M, Taatjes DJ, Kovarik P. Bancerek J, et al. Immunity. 2013 Feb 21;38(2):250-62. doi: 10.1016/j.immuni.2012.10.017. Epub 2013 Jan 24. Immunity. 2013. PMID: 23352233 Free PMC article.
References
- Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–20. - PubMed
- Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–7. - PubMed
- Vogelstein B, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. - PubMed
- van de Wetering M, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R33 CA128625/CA/NCI NIH HHS/United States
- K08 CA134931/CA/NCI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- T32 CA009172/CA/NCI NIH HHS/United States
- R33 CA128625-01A1/CA/NCI NIH HHS/United States
- P50CA127003/CA/NCI NIH HHS/United States
- R33CA128625/CA/NCI NIH HHS/United States
- P50 CA127003/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials