Delivery of steric block morpholino oligomers by (R-X-R)4 peptides: structure-activity studies - PubMed (original) (raw)
Comparative Study
. 2008 Nov;36(20):6343-54.
doi: 10.1093/nar/gkn541. Epub 2008 Sep 16.
Affiliations
- PMID: 18796528
- PMCID: PMC2582615
- DOI: 10.1093/nar/gkn541
Comparative Study
Delivery of steric block morpholino oligomers by (R-X-R)4 peptides: structure-activity studies
Rachida Abes et al. Nucleic Acids Res. 2008 Nov.
Abstract
Redirecting the splicing machinery through the hybridization of high affinity, RNase H- incompetent oligonucleotide analogs such as phosphoramidate morpholino oligonucleotides (PMO) might lead to important clinical applications. Chemical conjugation of PMO to arginine-rich cell penetrating peptides (CPP) such as (R-Ahx-R)(4) (with Ahx standing for 6-aminohexanoic acid) leads to sequence-specific splicing correction in the absence of endosomolytic agents in cell culture at variance with most conventional CPPs. Importantly, (R-Ahx-R)(4)-PMO conjugates are effective in mouse models of various viral infections and Duchenne muscular dystrophy. Unfortunately, active doses in some applications might be close to cytotoxic ones thus presenting challenge for systemic administration of the conjugates in those clinical settings. Structure-activity relationship studies have thus been undertaken to unravel CPP structural features important for the efficient nuclear delivery of the conjugated PMO and limiting steps in their internalization pathway. Affinity for heparin (taken as a model heparan sulfate), hydrophobicity, cellular uptake, intracellular distribution and splicing correction have been monitored. Spacing between the charges, hydrophobicity of the linker between the Arg-groups and Arg-stereochemistry influence splicing correction efficiency. A significant correlation between splicing correction efficiency, affinity for heparin and ability to destabilize model synthetic vesicles has been observed but no correlation with cellular uptake has been found. Efforts will have to focus on endosomal escape since it appears to remain the limiting factor for the delivery of these splice-redirecting ON analogs.
Figures
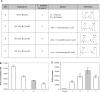
Figure 1.
Effect of charge spacing of (R-X-R)_n_–PMO conjugates. (A) Structures and nomenclature. (B) Heparin affinity chromatography. (R-X-R)_n_–PMO conjugates (white bars) and the reference (R-Ahx-R)4–PMO (gray bar) were injected on a HiTrap Sepharose/heparin column and eluted by a linear gradient of NaCl. Elution was monitored by UV absorption at 260 nm. Results are presented as eluting NaCl concentration. Each experiment was made in triplicate. Means of triplicates and standard deviations (error bars) are indicated. Mean values for all conjugates were compared to the mean value of the reference conjugate using Student's _t_-test (*** and ** indicate statistically significant differences; NS indicates that the difference is not statistically significant). (C) Splicing correction efficiency. HeLa pLuc705 cells were incubated for 4 h in OptiMEM in the presence of 1 μM (white bars) of the various (R-X-R)_n_–PMO conjugates or with 1 μM of the (R-Ahx-R)4–PMO reference compound (gray bars). Luciferase expression was quantified 20 h later and expressed as RLU per microgram protein. Each experiment was made in triplicate. Mean values and standard deviations (error bars) are indicated. Mean values for all conjugates were compared to the mean value of the reference conjugate using Student's _t_-test. 1, (R-G-R)4–PMO; 3, (R-Abu-R)4–PMO; 5, (R-Ahx-R)4–PMO; 7, (R-Acy-R)4–PMO.
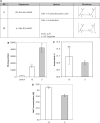
Figure 2.
Effect of hydrophobicity of the linker in (R-X-R)_n_–PMO conjugates. (A) Structures and nomenclature. (B) Hydrophobicity of (R-X-R)_n_–PMO conjugates. (R-X-R)_n_–PMO conjugates were injected on a C18-Sepharose column and eluted by a linear gradient of acetonitrile. Elution was monitored by UV absorption at 260 nm. Results are presented as eluting acetonitrile concentrations. Each experiment was made in triplicate. Means of triplicates and standard deviations (error bars) are indicated. Mean values for all conjugates were compared to the mean value of the (R-Ahx-R)4–PMO reference conjugate using Student's _t_-test. 7, (R-Acy-R)4–PMO; 8, (R-AbuF-R)4–PMO; 9, (R-AbuL-R)4–PMO; 10, (R-AbuNLe-R)4–PMO; 11, (R-AbuA-R)4–PMO; 5, (R-Ahx- R)4–PMO. (C) Splicing correction efficiency. Cells were treated and data were processed as described in the legend of Figure 1C. (D) Heparin affinity chromatography. Samples were treated and data were processed as described in the legend of Figure 1B. *** and ** Indicate statistically significant differences; NS indicates that the difference is not statistically significant.
Figure 3.
Effect of stereochemistry. (A) Structures and nomenclature. (B) Splicing correction efficiency. Cells were treated and data were processed as described in the legend of Figure 1C. 5, (R-Ahx-R)4–PMO; 13, (r-Ahx- R)4–PMO. (C) Hydrophobicity chromatography. Samples were treated and data were processed as described in the legend of Figure 2B. (D) Heparin affinity chromatography. Samples were treated and data were processed as described in the legend of Figure 1B. *** and ** Indicate statistically significant differences; NS indicates that the difference is not statistically significant.
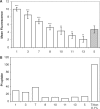
Figure 4.
Flow cytometry analysis. (A) Cell uptake of the FAM-labeled (R-X-R)4–PMO conjugates. HeLa pLuc705 cells were incubated for 1 h in Opti MEM with 1 μM of the various FAM-labeled (R-X-R)4–PMO conjugates (white bars) or with 1 μM of the FAM-labeled (R-Ahx-R)4–PMO reference compound (gray bar). Cells were washed, trypsinized and analyzed by flow cytometry. Each experiment was made in triplicate. Means of triplicates and standard deviations (error bars) are indicated. Mean values for all conjugates were compared to the mean value of the (R-Ahx-R)4–PMO reference conjugate using Student's _t_-test. See Figures 1A, 2A and 3A for structures. (B) PI uptake in (R-X-R)4–PMO conjugate-treated cells. (R-X-R)4–PMO conjugate-treated cells were incubated with PI immediately before FACS analysis. Data were processed as described in A.
Figure 5.
Effect of the number of (R-X-R) repeats onj splicing correction. HeLa pLuc705 cells were incubated for 4 h in Opti MEM with 1 μM of (R-Ahx-R)4–PMO, (R-AbuL-R)4–PMO or (R-AbuL-R)3–PMO. Luciferase expression was quantified 20 h later and expressed as RLU per microgram protein. Each experiment was made in triplicate. Means of triplicates and standard deviations (error bars) are indicated. Mean values for all conjugates were compared to the mean value of the (R-Ahx-R)4–PMO reference conjugate using Student's _t_-test.
Figure 6.
Effect of saponin on the intracellular distribution of FAM-labeled (R-Ahx-R)_n_–PMO and Alexa-labeled transferrin. HeLa pLuc 705 cells were incubated in OptiMEM with 2 µM of (R-Ahx-R)4–PMO–FAM in the absence (A) or in the presence of saponin (B) for 30 min. Hoechst dye blue and Alexa-labeled transferrin were then added for 10 min as markers for cell nuclei (blue fluorescence) and for endosomes (red fluorescence), respectively. Live unfixed cells were analyzed by fluorescence microscopy. Filter selection allows the detection of transferrin (A1 and B1) or of (R-Ahx-R)4–PMO (A2 and B2). In C, cells have been incubated for 30 min with Alexa-labeled transferrin and either observed immediately (C1) or after a 30 min treatment with saponin (C2).
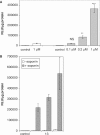
Figure 7.
Luciferase expression in saponin-permeabilized cells. (A) Splicing correction in dose–response. HeLa pLuc 705 cells were incubated for 30 min in OptiMEM at the indicated concentrations of the (R-Ahx-R)4–PMO conjugate in the absence (white bar) or in the presence of 20 µg/ml saponin (gray bars). Luciferase expression was quantified 24 h later and expressed as RLU per microgram protein. Each experiment was made in triplicate. Means of triplicates and standard deviations (error bars) are indicated. Mean values for all conjugates were compared to the mean value of the (R-Ahx-R)4–PMO reference conjugate using Student's _t_-test. (B) Splicing correction efficiency by various (R-X-R)_n_–PMO conjugates. HeLa pLuc 705 cells were incubated for 30 min in OptiMEM with 1 μM of the conjugates in the absence (white bars; −) or in the presence of 20 μg/ml saponin (gray bars; +). Luciferase expression was quantified 24 h later and expressed as RLU per microgram protein. Each experiment was made in triplicate. Means of triplicates and standard deviations (error bars) are indicated. Mean values for all conjugates were compared to the mean value of the (R-Ahx-R)4–PMO reference conjugate using Student's _t_-test. 1, (R-G-R)4–PMO; 13, (r-Ahx-R)4–PMO; 5, (R-Ahx-R)4–PMO.
Figure 8.
CPP–PMO-induced leakage of encapsulated dye from lipid vesicles mimicking late endosome membrane. (A) Influence of CPP structure. Conjugates (5 μM final concentration) were added to 2 ml of liposomes (25 μM) loaded with ANTS and DPX in MES buffer (pH 5.5). Single representative curves are shown for each conjugate. (R-G-R)4–PMO (black curve); (R-Ahx-R)4–PMO (red curve); (R-AbuL-R)4–PMO (green curve); R8-PMO (yellow curve). (B) pH dependence. Extent of encapsulated dye leakage after 30 min incubation with different CPP–PMO conjugates was measured in either Tris (pH 7.4: white bars) or MES (pH 5.5: black bars) buffer. Each experiment was made in triplicate. Mean and standard deviation (error bars) are shown. Increase in leakage extent at pH 5.5 as compared to pH 7 is statistically significant for conjugates 1 and 5. At pH 5.5 differences in leakage extents between all tested conjugates are statistically significantly. Statistical significance of the difference in the mean values was tested using paired, two-tailed Student's _t_-test at significance level of 0.05. 1, (R-G-R)4–PMO; 5, (R-Ahx-R)4–PMO; 9, (R-AbuL-R)4–PMO.
Similar articles
- Arginine-rich cell penetrating peptides: design, structure-activity, and applications to alter pre-mRNA splicing by steric-block oligonucleotides.
Abes R, Arzumanov A, Moulton H, Abes S, Ivanova G, Gait MJ, Iversen P, Lebleu B. Abes R, et al. J Pept Sci. 2008 Apr;14(4):455-60. doi: 10.1002/psc.979. J Pept Sci. 2008. PMID: 18236382 Review. - Vectorization of morpholino oligomers by the (R-Ahx-R)4 peptide allows efficient splicing correction in the absence of endosomolytic agents.
Abes S, Moulton HM, Clair P, Prevot P, Youngblood DS, Wu RP, Iversen PL, Lebleu B. Abes S, et al. J Control Release. 2006 Dec 1;116(3):304-13. doi: 10.1016/j.jconrel.2006.09.011. Epub 2006 Sep 30. J Control Release. 2006. PMID: 17097177 - Cell penetrating peptide conjugates of steric block oligonucleotides.
Lebleu B, Moulton HM, Abes R, Ivanova GD, Abes S, Stein DA, Iversen PL, Arzumanov AA, Gait MJ. Lebleu B, et al. Adv Drug Deliv Rev. 2008 Mar 1;60(4-5):517-29. doi: 10.1016/j.addr.2007.09.002. Epub 2007 Oct 22. Adv Drug Deliv Rev. 2008. PMID: 18037527 Free PMC article. Review. - Cell-penetrating peptides as transporters for morpholino oligomers: effects of amino acid composition on intracellular delivery and cytotoxicity.
Wu RP, Youngblood DS, Hassinger JN, Lovejoy CE, Nelson MH, Iversen PL, Moulton HM. Wu RP, et al. Nucleic Acids Res. 2007;35(15):5182-91. doi: 10.1093/nar/gkm478. Epub 2007 Aug 1. Nucleic Acids Res. 2007. PMID: 17670797 Free PMC article. - Cell-penetrating-peptide-based delivery of oligonucleotides: an overview.
Abes R, Arzumanov AA, Moulton HM, Abes S, Ivanova GD, Iversen PL, Gait MJ, Lebleu B. Abes R, et al. Biochem Soc Trans. 2007 Aug;35(Pt 4):775-9. doi: 10.1042/BST0350775. Biochem Soc Trans. 2007. PMID: 17635146 Review.
Cited by
- Cell penetrating peptides: overview and applications to the delivery of oligonucleotides.
Said Hassane F, Saleh AF, Abes R, Gait MJ, Lebleu B. Said Hassane F, et al. Cell Mol Life Sci. 2010 Mar;67(5):715-26. doi: 10.1007/s00018-009-0186-0. Epub 2009 Nov 7. Cell Mol Life Sci. 2010. PMID: 19898741 Free PMC article. Review. - The chemistry and biology of oligonucleotide conjugates.
Juliano RL, Ming X, Nakagawa O. Juliano RL, et al. Acc Chem Res. 2012 Jul 17;45(7):1067-76. doi: 10.1021/ar2002123. Epub 2012 Feb 21. Acc Chem Res. 2012. PMID: 22353142 Free PMC article. - Synthesis and splice-redirecting activity of branched, arginine-rich peptide dendrimer conjugates of peptide nucleic acid oligonucleotides.
Saleh AF, Arzumanov A, Abes R, Owen D, Lebleu B, Gait MJ. Saleh AF, et al. Bioconjug Chem. 2010 Oct 20;21(10):1902-11. doi: 10.1021/bc100275r. Bioconjug Chem. 2010. PMID: 20879728 Free PMC article. - Generation of oligonucleotide conjugates via one-pot diselenide-selenoester ligation-deselenization/alkylation.
Liczner C, Hanna CC, Payne RJ, Wilds CJ. Liczner C, et al. Chem Sci. 2021 Nov 19;13(2):410-420. doi: 10.1039/d1sc04937b. eCollection 2022 Jan 5. Chem Sci. 2021. PMID: 35126973 Free PMC article. - Inhibition of SARS-CoV-2 growth in the lungs of mice by a peptide-conjugated morpholino oligomer targeting viral RNA.
Sakai A, Singh G, Khoshbakht M, Bittner S, Löhr CV, Diaz-Tapia R, Warang P, White K, Luo LL, Tolbert B, Blanco M, Chow A, Guttman M, Li C, Bao Y, Ho J, Maurer-Stroh S, Chatterjee A, Chanda S, García-Sastre A, Schotsaert M, Teijaro JR, Moulton HM, Stein DA. Sakai A, et al. Mol Ther Nucleic Acids. 2024 Sep 10;35(4):102331. doi: 10.1016/j.omtn.2024.102331. eCollection 2024 Dec 10. Mol Ther Nucleic Acids. 2024. PMID: 39376996 Free PMC article.
References
- Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait JM, Chernomordik VL, Lebleu B. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J. Biol. Chem. 2003;278:585–590. - PubMed
- Abes S, Moulton MH, Clair P, Prevot P, Youngblood SD, Wu PR, Iversen LP, Lebleu B. Vectorization of morpholino oligomers by the (R-Ahx-R)4 peptide allows efficient splicing correction in the absence of endosomolytic agents. J. Control Release. 2006;116:304–313. - PubMed
- Moulton HM, Nelson HM, Hatlevig AS, Reddy TM, Iversen LP. Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjug. Chem. 2004;15:290–299. - PubMed
- McClorey G, Moulton MH, Iversen LP, Fletcher S, Wilton DS. Antisense oligonucleotide-induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Ther. 2006;13:1373–1381. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous