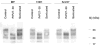The effects of prion protein proteolysis and disaggregation on the strain properties of hamster scrapie - PubMed (original) (raw)
The effects of prion protein proteolysis and disaggregation on the strain properties of hamster scrapie
Andrea M Deleault et al. J Gen Virol. 2008 Oct.
Abstract
Native mammalian prions exist in self-propagating strains that exhibit distinctive clinical, pathological and biochemical characteristics. Prion strain diversity is associated with variations in PrP(Sc) conformation, but it remains unknown precisely which physical properties of the PrP(Sc) molecules are required to encipher mammalian prion strain phenotypes. In this study, we subjected prion-infected brain homogenates derived from three different hamster scrapie strains to either (i) proteinase K digestion or (ii) sonication, and inoculated the modified samples into normal hamsters. The results show that the strain-specific clinical features and neuropathological profiles of inoculated animals were not affected by either treatment. Similarly, the strain-dependent biochemical characteristics of the PrP(Sc) molecules (including electrophoretic mobility, glycoform composition, conformational stability and susceptibility to protease digestion) in infected animals were unaffected by either proteolysis or sonication of the original inocula. These results indicate that the infectious strain properties of native prions do not appear to be altered by PrP(Sc) disaggregation, and that maintenance of such properties does not require the N-domain (approximately residues 23-90) of the protease-resistant PrP(Sc) molecules or protease-sensitive PrP(Sc) molecules.
Figures
Fig. 1
Western blot analysis of brain homogenate samples inoculated into Syrian hamsters. Three different scrapie strains were used to generate samples: DY, 139H and Sc237. Control, PrP27–30 and sonicated samples were generated as described in Methods.
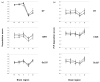
Fig. 2
Neuropathology of scrapie-infected hamsters. (a) Vacuolation profile scores of animals inoculated with samples derived from different prion strains, as indicated. (b) PrP immunohistochemistry profiles of animals inoculated with samples derived from different prion strains, as indicated. (a and b) Symbols: black squares, animals inoculated with control scrapie brain homogenate; open circles, animals inoculated with PrP27–30 samples; grey triangles, animals inoculated with sonicated samples. Brain regions: FC, frontal cortex; PC, parietal cortex; H, hippocampus; C, cerebellum; M, medulla. The mean values (_n_=5–8 animals per group) are shown ± SEM.
Fig. 3
Biochemical analyses of scrapie-infected hamster brains. (a) Western blot analysis of brain homogenate samples prepared from animals inoculated with samples derived from different prion strains, as indicated. Twenty-five microlitres was loaded for each sample not treated with PK (−), and 50 μl was loaded for each sample subjected to digestion with 50 μg PK ml−1 (+). (b) Western blot analysis of brain homogenate samples prepared from animals inoculated with samples derived from different prion strains, as indicated. All samples were incubated with 10 μg PK ml−1 for 30 min at 37 _°_C. Samples on the right side of the blot were treated with the enzyme PNGase F, as indicated.
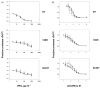
Fig. 4
Biochemical analysis of PrPSc stability. (a) Protease resistance assay showing PrPSc levels in samples of brain homogenates taken from animals inoculated with samples derived from different prion strains, as indicated, and incubated with various concentrations of PK as described in Methods. (b) Guanidine denaturation assay showing PrPSc levels in samples of brain homogenates taken from animals inoculated with samples derived from different prion strains, as indicated, and incubated with various concentrations of guanidine hydrochloride, as described in Methods. (a and b) Symbols: black squares, animals inoculated with control scrapie brain homogenate; open circles, animals inoculated with PrP27–30 samples; grey triangles, animals inoculated with sonicated samples. The mean values of replicates (_n_=3) are shown for each point ± SEM.
References
- Barron RM, Campbell SL, King D, Bellon A, Chapman KE, Williamson RA, Manson JC. High titers of transmissible spongiform encephalopathy infectivity associated with extremely low levels of PrPScin vivo. J Biol Chem. 2007;282:35878–35886. - PubMed
- Brown P, Gajdusek DC. Survival of scrapie virus after 3 years’ interment. Lancet. 1991;337:269–270. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials