Updated three-stage model for the peopling of the Americas - PubMed (original) (raw)
Updated three-stage model for the peopling of the Americas
Connie J Mulligan et al. PLoS One. 2008.
Abstract
Background: We re-assess support for our three stage model for the peopling of the Americas in light of a recent report that identified nine non-Native American mitochondrial genome sequences that should not have been included in our initial analysis. Removal of these sequences results in the elimination of an early (i.e. approximately 40,000 years ago) expansion signal we had proposed for the proto-Amerind population.
Methodology/findings: Bayesian skyline plot analysis of a new dataset of Native American mitochondrial coding genomes confirms the absence of an early expansion signal for the proto-Amerind population and allows us to reduce the variation around our estimate of the New World founder population size. In addition, genetic variants that define New World founder haplogroups are used to estimate the amount of time required between divergence of proto-Amerinds from the Asian gene pool and expansion into the New World.
Conclusions/significance: The period of population isolation required for the generation of New World mitochondrial founder haplogroup-defining genetic variants makes the existence of three stages of colonization a logical conclusion. Thus, our three stage model remains an important and useful working hypothesis for researchers interested in the peopling of the Americas and the processes of colonization.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
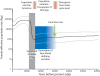
Figure 1. Bayesian skyline plot of 148 Native American mitochondrial coding genome sequences.
The curve plots median Nef with 95% credible intervals indicated by light gray lines. The shaded gray box highlights the significant increase of Nef during the colonization of the Americas 16–12 kya. The blue box depicts the calculated time required for the generation of New World defining mitochondrial variants and its shaded region represents the variation in these estimates, i.e. 7–15 thousand years before entry to the New World (see Table 1). The green arrow identifies the date of the Yana River site of human occupation in western Beringia .
Similar articles
- A three-stage colonization model for the peopling of the Americas.
Kitchen A, Miyamoto MM, Mulligan CJ. Kitchen A, et al. PLoS One. 2008 Feb 13;3(2):e1596. doi: 10.1371/journal.pone.0001596. PLoS One. 2008. PMID: 18270583 Free PMC article. - A reevaluation of the Native American mtDNA genome diversity and its bearing on the models of early colonization of Beringia.
Fagundes NJ, Kanitz R, Bonatto SL. Fagundes NJ, et al. PLoS One. 2008 Sep 17;3(9):e3157. doi: 10.1371/journal.pone.0003157. PLoS One. 2008. PMID: 18797501 Free PMC article. - Mitochondrial echoes of first settlement and genetic continuity in El Salvador.
Salas A, Lovo-Gómez J, Alvarez-Iglesias V, Cerezo M, Lareu MV, Macaulay V, Richards MB, Carracedo A. Salas A, et al. PLoS One. 2009 Sep 2;4(9):e6882. doi: 10.1371/journal.pone.0006882. PLoS One. 2009. PMID: 19724647 Free PMC article. - Population genetics, history, and health patterns in native americans.
Mulligan CJ, Hunley K, Cole S, Long JC. Mulligan CJ, et al. Annu Rev Genomics Hum Genet. 2004;5:295-315. doi: 10.1146/annurev.genom.5.061903.175920. Annu Rev Genomics Hum Genet. 2004. PMID: 15485351 Review. - The late Pleistocene dispersal of modern humans in the Americas.
Goebel T, Waters MR, O'Rourke DH. Goebel T, et al. Science. 2008 Mar 14;319(5869):1497-502. doi: 10.1126/science.1153569. Science. 2008. PMID: 18339930 Review.
Cited by
- Bidirectional dispersals during the peopling of the North American Arctic.
Luis JR, Palencia-Madrid L, Garcia-Bertrand R, Herrera RJ. Luis JR, et al. Sci Rep. 2023 Jan 23;13(1):1268. doi: 10.1038/s41598-023-28384-8. Sci Rep. 2023. PMID: 36690673 Free PMC article. - Beringia and the peopling of the Western Hemisphere.
Hoffecker JF, Elias SA, Scott GR, O'Rourke DH, Hlusko LJ, Potapova O, Pitulko V, Pavlova E, Bourgeon L, Vachula RS. Hoffecker JF, et al. Proc Biol Sci. 2023 Jan 11;290(1990):20222246. doi: 10.1098/rspb.2022.2246. Epub 2023 Jan 11. Proc Biol Sci. 2023. PMID: 36629115 Free PMC article. Review. - A genomic perspective on South American human history.
Silva MACE, Ferraz T, Hünemeier T. Silva MACE, et al. Genet Mol Biol. 2022 Jul 29;45(3 Suppl 1):e20220078. doi: 10.1590/1678-4685-GMB-2022-0078. eCollection 2022. Genet Mol Biol. 2022. PMID: 35925590 Free PMC article. - Novel alleles gained during the Beringian isolation period.
Niedbalski SD, Long JC. Niedbalski SD, et al. Sci Rep. 2022 Mar 11;12(1):4289. doi: 10.1038/s41598-022-08212-1. Sci Rep. 2022. PMID: 35277570 Free PMC article. - Moose genomes reveal past glacial demography and the origin of modern lineages.
Dussex N, Alberti F, Heino MT, Olsen RA, van der Valk T, Ryman N, Laikre L, Ahlgren H, Askeyev IV, Askeyev OV, Shaymuratova DN, Askeyev AO, Döppes D, Friedrich R, Lindauer S, Rosendahl W, Aspi J, Hofreiter M, Lidén K, Dalén L, Díez-Del-Molino D. Dussex N, et al. BMC Genomics. 2020 Dec 2;21(1):854. doi: 10.1186/s12864-020-07208-3. BMC Genomics. 2020. PMID: 33267779 Free PMC article.
References
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. - PubMed
- Bandelt HJ, Herrnstadt C, Yao YG, Kong QP, Kivisild T, et al. Identification of Native American founder mtDNAs through the analysis of complete mtDNA sequences: some caveats. Ann Hum Genet. 2003;67:512–524. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources