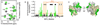Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12 - PubMed (original) (raw)
Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12
Christopher T Veldkamp et al. Sci Signal. 2008.
Abstract
Stem cell homing and breast cancer metastasis are orchestrated by the chemokine stromal cell-derived factor 1 (SDF-1) and its receptor CXCR4. Here, we report the nuclear magnetic resonance structure of a constitutively dimeric SDF-1 in complex with a CXCR4 fragment that contains three sulfotyrosine residues important for a high-affinity ligand-receptor interaction. CXCR4 bridged the SDF-1 dimer interface so that sulfotyrosines sTyr7 and sTyr12 of CXCR4 occupied positively charged clefts on opposing chemokine subunits. Dimeric SDF-1 induced intracellular Ca2+ mobilization but had no chemotactic activity; instead, it prevented native SDF-1-induced chemotaxis, suggesting that it acted as a potent partial agonist. Our work elucidates the structural basis for sulfotyrosine recognition in the chemokine-receptor interaction and suggests a strategy for CXCR4-targeted drug development.
Figures
Fig. 1
The NMR structure of disulfide-locked SDF12. (A) The amino acid sequence of SDF12 with the conserved intramolecular disulfide bonds (black lines) and the engineered intermolecular disulfide bonds (red lines) illustrated. (B) SDS-PAGE of SDF-1 and SDF12 treated with or without dithiothreitol (DTT). SDF-1 and SDF12 migrate near the monomeric molecular weight of 8 kD when treated with DTT. In contrast, whereas SDF12 migrates as a dimer, SDF-1 migrates as a monomer in the absence of DTT. (C) Translational diffusion measurements of SDF12 indicate that SDF12 is dimeric. Diffusion coefficients (Ds) of wild-type SDF-1 (black circles) in 20 mM sodium phosphate at pH 7.4 plotted against chemokine concentration (22). Nonlinear fitting of the Ds values of SDF-1 indicates a dimer dissociation Kd of 120 µM with a pure monomer Ds value of ~ 1.6 (×10−6 cm2s−1) and a dimer value of ~1.0 (×10−6 cm2s−1) [data from Veldkamp et al. (22)]. Ds values for 10, 50, and 150 µM SDF12 (red triangles) range from 1.08–1.09 (×10−6 cm2s−1) consistent with those expected for SDF-1 in the dimeric state (data from this study). (D) Ensemble of 20 NMR solution structures of SDF12 (gray and tan) superimposed on the crystal structure of dimeric wild-type SDF-1 (blue, PDB ID 2J7Z) with an α-carbon RMSD of 1.2 Å for residues 9–66. Intermolecular Cys36-Cys65 disulfide bonds are shown in yellow. Flexible N-terminal residues of SDF-1 (–8) are omitted for clarity. Refinement statistics for the SDF12 structure ensemble are given in table S1.
Fig. 2
The N-terminus of CXCR4 binds to SDF12. (A) 15N-1H HSQC spectra of 25 µM [U-15N]-SDF12 alone (black contours) and after the addition of 100 µM p38 peptide (green contours). (B) Combined 15N-1H chemical shift perturbations plotted against SDF12 residue number. Secondary structure elements are indicated and regions involved in the dimer interface are highlighted in orange. Missing values correspond to proline residues (sequence positions , , , and 53) or amino acid residues not observed in the 15N-1H HSQC spectra. (C) Chemical shift mapping on the SDF12 structure. Green surface highlighting corresponds to shift perturbations > 0.25 in (B).
Fig. 3
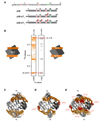
Structures of SDF12 dimers bound to the N-terminal domain of CXCR4. (A) N-terminal peptides corresponding to the first 38 amino acids of CXCR4 are illustrated. The sequence for p38 is identical to that of CXCR4 except for a Gly-Ser dipeptide on the N-terminus, which results from a cloning artifact, and a Cys28 → Ala28 mutation to prevent oxidative peptide dimer formation. The sulfated peptides are identical to p38 except for the inclusion of sulfotyrosine at position 21 for p38-sY1 and at 7, 12, and 21 for p38-sY3. (B) Representative intermolecular NOEs for the SDF12:p38-sY1 complex. Strips from 3D F1-13C-fliltered/F3-13C-edited NOESY-HSQC spectra acquired from a complex containing [U-15N,13C]-SDF12 and unlabeled p38-sY1 (left) and a complex containing [U-15N,13C]-p38-sY1 and unlabeled SDF12 (right) contain equivalent NOEs between the methyl group of Val18 of SDF12 and sTyr21 1Hδ of p38-sY1. Ensembles of the 20 lowest energy conformers for the SDF12:p38 (C), SDF12:p38-sY1 (D), and SDF12:p38-sY3 (E) complexes. SDF12 is shown in gray and the CXCR4 N-termini are orange. Sulfotyrosine residues in N-termini of CXCR4 are shown in red.
Fig. 4
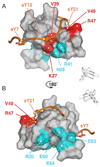
Recognition of sulfotyrosines by SDF12. (A) NMR structure of SDF12 bound to p38-sY3. Individual subunits of the symmetric SDF12 dimer are shown in tan and white with symmetry-related p38-sY3 peptides in blue and orange. Chemical shift perturbations greater than 0.25 ppm (Fig. 2C) are highlighted in green on the surface of SDF12. Flexible regions of SDF12 (residues –8) and p38-sY3 (residues –38) are omitted for clarity. Sulfotyrosine side chains are shown in a ball-and-stick representation. In panels B–D, basic residues in SDF12 that pair with CXCR4 sulfotyrosines are shown in blue and SDF12 residues with NOEs to the sulfotyrosines are shown in green. (B) The sTyr7 residue of CXCR4 binds to SDF12 near Arg20 and makes NOE contacts with Val23. (C) The sTyr12 residue of CXCR4 occupies a cleft bounded by residues Lys27, Pro10, and Leu29 of SDF12. (D) The sTyr21 residue of CXCR4 pairs with Arg47 of SDF12 and makes NOE contacts with Val18 and Val49.
Fig. 5
Amino acid substitutions in native SDF-1 corroborate the CXCR4 N-terminal binding site. One subunit of the SDF12 dimer and one p38-sY3 molecule from the SDF12:p38-sY3 complex solved by NMR represent a model for the equivalent 1:1 complex. Front (A) and back (B) views of the SDF-1 surface are highlighted to indicate the location and functional impact of amino acid substitutions in the wild-type SDF-1 sequence. Substitutions at the sTyr12- and sTyr21-binding sites (red) showed increased EC50 values for Ca2+ mobilization, whereas substitutions away from the CXCR4-binding site (cyan) showed no change in their EC50 values. A binding site for sTyr7 is not defined in this model because sTyr7 binds to the opposing SDF-1 subunit in the SDF12:p38-sY3 structure.
Fig. 6
Dimeric SDF12 induces CXCR4-mediated Ca2+ mobilization but inhibits chemotaxis to wild-type SDF-1. (A) Ca2+ mobilization in THP-1 cells loaded with Fluo-3 indicates robust dose-dependant activation of CXCR4 by wild-type SDF-1 (●, EC50 = 3.6 nM) and SDF12 (▲, EC50 = 12.9 nM) (Data from Veldkamp et al. (53) and this study, respectively). (B) Wild-type SDF-1 induces the chemotaxis of THP-1 cells in a biphasic, concentration-dependent manner with a maximal migratory response at ~30 nM SDF-1. In contrast, SDF12 does not induce chemotaxis of THP-1 cells at any concentration from 1–1,000 nM. (C) Chemotaxis of THP-1 cells induced by 10 nM wild-type SDF-1 is inhibited by SDF12 (IC50 ~ 4 nM). (D) Wild-type SDF-1 and the dimerization-impaired His25 → Arg25 variant [SDF1(H25R)] induce chemotaxis of THP-1 cells equally well at low concentrations (0.1–10 nM). SDF1(H25R) remains monomeric at higher concentrations than does wild-type SDF-1 and induces chemotaxis over a broader range of concentrations. (E) Monomeric SDF-1 generates the full range of cellular responses to CXCR4 activation, whereas dimeric SDF1 is a partial agonist of CXCR4 that fails to induce chemotaxis. Loss of migration could be a consequence of aberrant CXCR4 trafficking.
Similar articles
- Recognition of a CXCR4 sulfotyrosine by the chemokine stromal cell-derived factor-1alpha (SDF-1alpha/CXCL12).
Veldkamp CT, Seibert C, Peterson FC, Sakmar TP, Volkman BF. Veldkamp CT, et al. J Mol Biol. 2006 Jun 23;359(5):1400-9. doi: 10.1016/j.jmb.2006.04.052. Epub 2006 May 11. J Mol Biol. 2006. PMID: 16725153 Free PMC article. - The role of tyrosine sulfation in the dimerization of the CXCR4:SDF-1 complex.
Rapp C, Snow S, Laufer T, McClendon CL. Rapp C, et al. Protein Sci. 2013 Aug;22(8):1025-36. doi: 10.1002/pro.2288. Epub 2013 Jun 29. Protein Sci. 2013. PMID: 23740770 Free PMC article. - Sulfopeptide probes of the CXCR4/CXCL12 interface reveal oligomer-specific contacts and chemokine allostery.
Ziarek JJ, Getschman AE, Butler SJ, Taleski D, Stephens B, Kufareva I, Handel TM, Payne RJ, Volkman BF. Ziarek JJ, et al. ACS Chem Biol. 2013 Sep 20;8(9):1955-63. doi: 10.1021/cb400274z. Epub 2013 Jun 26. ACS Chem Biol. 2013. PMID: 23802178 Free PMC article. - The good and bad faces of the CXCR4 chemokine receptor.
Teixidó J, Martínez-Moreno M, Díaz-Martínez M, Sevilla-Movilla S. Teixidó J, et al. Int J Biochem Cell Biol. 2018 Feb;95:121-131. doi: 10.1016/j.biocel.2017.12.018. Epub 2017 Dec 27. Int J Biochem Cell Biol. 2018. PMID: 29288743 Review. - Peptide and peptidomimetic ligands for CXC chemokine receptor 4 (CXCR4).
Oishi S, Fujii N. Oishi S, et al. Org Biomol Chem. 2012 Aug 14;10(30):5720-31. doi: 10.1039/c2ob25107h. Epub 2012 Apr 19. Org Biomol Chem. 2012. PMID: 22517031 Review.
Cited by
- A new obligate CXCL4-CXCL12 heterodimer for studying chemokine heterodimer activities and mechanisms.
Nguyen KTP, Volkman B, Dréau D, Nesmelova IV. Nguyen KTP, et al. Sci Rep. 2022 Oct 13;12(1):17204. doi: 10.1038/s41598-022-21651-0. Sci Rep. 2022. PMID: 36229490 Free PMC article. - Functional anatomy of the full-length CXCR4-CXCL12 complex systematically dissected by quantitative model-guided mutagenesis.
Stephens BS, Ngo T, Kufareva I, Handel TM. Stephens BS, et al. Sci Signal. 2020 Jul 14;13(640):eaay5024. doi: 10.1126/scisignal.aay5024. Sci Signal. 2020. PMID: 32665413 Free PMC article. - Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction.
Teramoto T, Fujikawa Y, Kawaguchi Y, Kurogi K, Soejima M, Adachi R, Nakanishi Y, Mishiro-Sato E, Liu MC, Sakakibara Y, Suiko M, Kimura M, Kakuta Y. Teramoto T, et al. Nat Commun. 2013;4:1572. doi: 10.1038/ncomms2593. Nat Commun. 2013. PMID: 23481380 Free PMC article. - Site-specific labeling of proteins with NMR-active unnatural amino acids.
Jones DH, Cellitti SE, Hao X, Zhang Q, Jahnz M, Summerer D, Schultz PG, Uno T, Geierstanger BH. Jones DH, et al. J Biomol NMR. 2010 Jan;46(1):89-100. doi: 10.1007/s10858-009-9365-4. Epub 2009 Aug 9. J Biomol NMR. 2010. PMID: 19669620 Review. - Evaluation and extension of the two-site, two-step model for binding and activation of the chemokine receptor CCR1.
Sanchez J, E Huma Z, Lane JR, Liu X, Bridgford JL, Payne RJ, Canals M, Stone MJ. Sanchez J, et al. J Biol Chem. 2019 Mar 8;294(10):3464-3475. doi: 10.1074/jbc.RA118.006535. Epub 2018 Dec 19. J Biol Chem. 2019. PMID: 30567735 Free PMC article.
References
- Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635. - PubMed
- Tachibana K, Hirota S, Iizasa H, Yoshida H, Kawabata K, Kataoka Y, Kitamura Y, Matsushima K, Yoshida N, Nishikawa S, Kishimoto T, Nagasawa T. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393:591. - PubMed
- Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595. - PubMed
- Bleul CC, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer TA. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829. - PubMed
- Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous