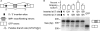Ab initio identification of functionally interacting pairs of cis-regulatory elements - PubMed (original) (raw)
Ab initio identification of functionally interacting pairs of cis-regulatory elements
Brad A Friedman et al. Genome Res. 2008 Oct.
Abstract
Cooperatively acting pairs of cis-regulatory elements play important roles in many biological processes. Here, we describe a statistical approach, compositionally orthogonalized co-occurrence analysis (coCOA) that detects pairs of oligonucleotides that preferentially co-occur in pairs of sequence regions, controlling for correlations between the compositions of the analyzed regions. coCOA identified three clusters of oligonucleotide pairs that frequently co-occur at 5' and 3' ends of human and mouse introns. The largest cluster involved GC-rich sequences at the 5' ends of introns that co-occur and are co-conserved with specific AU-rich sequences near intron 3' ends. These motifs are preferentially conserved when they occur together, as measured by a new co-conservation measure, supporting common in vivo function. These motif pairs are also enriched in introns flanking alternative "cassette" exons, suggesting a role in silencing of intervening exons, and we showed that these motifs can cooperatively silence splicing of an intervening exon in a splicing reporter assay. This approach can be easily generalized to problems beyond RNA splicing.
Figures
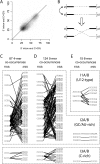
Figure 1.
coCOA detects three clusters of motif pairs that co-occur at 5′ and 3′ ends of human introns. (A) G+C content in the first 80 nt (_x_-axis) and last 80 nt (_y_-axis) of introns is correlated. A density plot of intron co-GC content is shown for a set of 53,326 constitutive human introns, with the darker/lighter squares corresponding to higher/lower intron density, respectively. The diagonal line y = x is shown for reference. (B) co-GC shuffling. (Above) Two hypothetical introns, A and B, with 5′/3′ ends a5/a3 and b5/b3. Intron A has high G+C content at both ends (thick lines). Intron B has high G+C content at the 5′ end, but lower G+C content near the 3′ end (thin solid line). Since the introns have similar G+C content at their 5′ ends, these ends can be swapped. (Below) Co-GC shuffled introns. The beginning of intron B (b5) is now paired with the end of intron A (a3), and the beginning of intron A (a5) is now paired with the end of intron B (b3). Overall co-GC content of the set of introns is preserved. (C–E) Preferentially co-occurring k_-mer pairs detected by coCOA are shown for k = 4, 5, and 6 at P ≤ 4−2_k, corresponding to a single expected false positive for each value of k. In each panel, _k_-mers occurring in the first 80 nt of introns are shown at left under “5′SS”; those occurring in the last 80 nt are shown at right under “3′SS”. The co-occurrences could all be grouped into three clusters, denoted I1, I2, and I3, with the 5′ss and 3′ss motifs designated A and B, respectively.
Figure 2.
Co-occurring motif pairs flanking alternative and constitutive exons and controls. Diagrams of the intron/exon data sets analyzed are shown at left, with exons shown as white boxes, introns as horizontal lines, and locations of the analyzed 80-nt regions shown as gray boxes. Splicing patterns are shown by angled lines; brackets indicate decoy splice sites. Representation of co-occurring _k_-mer pairs and _P_-value cutoffs as in Figure 1C–E. Numbers in parenthesis denote the number of significant _k_-mer pairs in each data set.
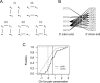
Figure 3.
The motif pair I2A/B co-occurs in mouse as well as human and is preferentially co-conserved. (A) Representation of five possible models for co-conservation. Lines represent dependencies that are modeled; absence of a line indicates assumption of independence. Model (v) is the maximum entropy model used to define the co-conservation rate. (B) The 23 HM-I2 tetramer pairs that significantly co-occur between the beginning/end of constitutive mouse as well as human introns are shown. (C) I2A/B motif pairs are more conserved than expected. Empirical cumulative distribution functions of chi statistic (higher values indicate increased conservation) for HM-I2 tetramer pairs and control pairs. Controls have similar numbers of co-occurrences in human and mouse constitutive introns as HM-I2 pairs. Vertical black and gray lines indicate mean of statistic over HM-I2 pairs and control pairs, respectively.
Figure 4.
I2A/B motif pairs can suppress splicing of an intervening exon. (A) Mini-gene construct for interrogating I2A/B motif pair, constructed by inserting exon 12 of the human IGF2BP1 gene and its flanking introns into the middle of the ORF in an eGFP expression construct. (B) The I2A/B motif pair promotes exon skipping in HeLa cells. The five indicated constructs containing I2A, I2B, or neutral (N) motifs inserted near the 5′ss or 3′ss were transfected into HeLa cells. Twenty-four hours later RNA was extracted and semi-quantitative RT-PCR using primers targeted to reporter exons 1 and 3 was performed to assay for relative isoform levels. (Top) Quantization of skipping levels. Data shown are mean + SEM for eight replicates—two PCRs for each of four transfection experiments. *I2A/B motif pair shows significantly more skipping than N/N control (P = 1.1 × 10−5 by one-sided _t_-test; 1.75-fold increase in skipping). At the 5% level, none of the other skipping levels is significantly greater than that of N/N. (Bottom) Representative gel showing levels of inclusion isoform (upper band) and skipping isoform (lower band). Last lane, intronless GFP control.
Similar articles
- Intronic motif pairs cooperate across exons to promote pre-mRNA splicing.
Ke S, Chasin LA. Ke S, et al. Genome Biol. 2010;11(8):R84. doi: 10.1186/gb-2010-11-8-r84. Epub 2010 Aug 12. Genome Biol. 2010. PMID: 20704715 Free PMC article. - A correlation with exon expression approach to identify cis-regulatory elements for tissue-specific alternative splicing.
Das D, Clark TA, Schweitzer A, Yamamoto M, Marr H, Arribere J, Minovitsky S, Poliakov A, Dubchak I, Blume JE, Conboy JG. Das D, et al. Nucleic Acids Res. 2007;35(14):4845-57. doi: 10.1093/nar/gkm485. Epub 2007 Jul 10. Nucleic Acids Res. 2007. PMID: 17626050 Free PMC article. - Conserved sequence elements associated with exon skipping.
Miriami E, Margalit H, Sperling R. Miriami E, et al. Nucleic Acids Res. 2003 Apr 1;31(7):1974-83. doi: 10.1093/nar/gkg279. Nucleic Acids Res. 2003. PMID: 12655015 Free PMC article. - Unannotated splicing regulatory elements in deep intron space.
Conboy JG. Conboy JG. Wiley Interdiscip Rev RNA. 2021 Sep;12(5):e1656. doi: 10.1002/wrna.1656. Epub 2021 Apr 22. Wiley Interdiscip Rev RNA. 2021. PMID: 33887804 Review. - Searching for splicing motifs.
Chasin LA. Chasin LA. Adv Exp Med Biol. 2007;623:85-106. doi: 10.1007/978-0-387-77374-2_6. Adv Exp Med Biol. 2007. PMID: 18380342 Review.
Cited by
- A biophysical model for identifying splicing regulatory elements and their interactions.
Wen J, Chen Z, Cai X. Wen J, et al. PLoS One. 2013;8(1):e54885. doi: 10.1371/journal.pone.0054885. Epub 2013 Jan 30. PLoS One. 2013. PMID: 23382993 Free PMC article. - A network of conserved co-occurring motifs for the regulation of alternative splicing.
Suyama M, Harrington ED, Vinokourova S, von Knebel Doeberitz M, Ohara O, Bork P. Suyama M, et al. Nucleic Acids Res. 2010 Dec;38(22):7916-26. doi: 10.1093/nar/gkq705. Epub 2010 Aug 11. Nucleic Acids Res. 2010. PMID: 20702423 Free PMC article. - Intronic motif pairs cooperate across exons to promote pre-mRNA splicing.
Ke S, Chasin LA. Ke S, et al. Genome Biol. 2010;11(8):R84. doi: 10.1186/gb-2010-11-8-r84. Epub 2010 Aug 12. Genome Biol. 2010. PMID: 20704715 Free PMC article. - Context-dependent splicing regulation: exon definition, co-occurring motif pairs and tissue specificity.
Ke S, Chasin LA. Ke S, et al. RNA Biol. 2011 May-Jun;8(3):384-8. doi: 10.4161/rna.8.3.14458. Epub 2011 May 1. RNA Biol. 2011. PMID: 21444999 Free PMC article. - Networking in a global world: establishing functional connections between neural splicing regulators and their target transcripts.
Calarco JA, Zhen M, Blencowe BJ. Calarco JA, et al. RNA. 2011 May;17(5):775-91. doi: 10.1261/rna.2603911. Epub 2011 Mar 17. RNA. 2011. PMID: 21415141 Free PMC article. Review.
References
- Bailey T.L., Elkan C., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. - PubMed
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. - PubMed
- Burge C.B., Padgett R.A., Sharp P.A., Padgett R.A., Sharp P.A., Sharp P.A. Evolutionary fates and origins of U12-type introns. Mol. Cell. 1998;2:773–785. - PubMed
- Burnette J.M., Miyamoto-Sato E., Schaub M.A., Conklin J., Lopez A.J., Miyamoto-Sato E., Schaub M.A., Conklin J., Lopez A.J., Schaub M.A., Conklin J., Lopez A.J., Conklin J., Lopez A.J., Lopez A.J. Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics. 2005;170:661–674. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous