Chemical probes for histone-modifying enzymes - PubMed (original) (raw)
Review
Chemical probes for histone-modifying enzymes
Philip A Cole. Nat Chem Biol. 2008 Oct.
Abstract
The histone-modifying enzymes that catalyze reversible lysine acetylation and methylation are central to the epigenetic regulation of chromatin remodeling. From the early discovery of histone deacetylase inhibitors to the more recent identification of histone demethylase blockers, chemical approaches offer increasingly sophisticated tools for the investigation of the structure and function of these lysine-modifying enzymes. This review summarizes progress to date on compounds identified from screens or by design that can modulate the activity of classical histone deacetylases, sirtuins, histone acetyltransferases, histone methyltransferases and histone demethylases. We highlight applications of compounds to mechanistic and functional studies involving these enzymes and discuss future challenges regarding target specificity and general utility.
Figures
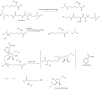
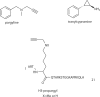
Figure 1
Reversible histone acetylation catalyzed by histone acetyltransferases (HATs), classical histone deacetylases (HDACs), and sirtuins (Sir2s). Transferred acetyl group is highlighted in blue. R = 3',5'-adenosine diphosphate; R1 = adenosine 5'-diphosphate.
Figure 2
Reversible histone methylation catalyzed by histone methyltransferases, LSD1 demethylase, and Jmj demethylases. Transferred methyl group highlighted in red. R = methyl or hydrogen; R1 = ribose-adenosine 5'-diphosphosphate.
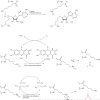
Figure 3
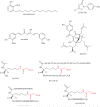
Selected classical histone deacetylase inhibitors. Warhead features highlighted in red.
Figure 4
Activators and inhibitors of Sir2 enzymes.
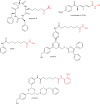
Figure 5
Natural product and synthetic histone acetyltransferase inhibitors.
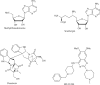
Fig. 6
Histone methyltransferase inhibitors.
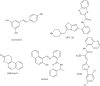
Figure 7
Histone demethylase inhibitors.
Similar articles
- Small molecule control of chromatin remodeling.
Finley A, Copeland RA. Finley A, et al. Chem Biol. 2014 Sep 18;21(9):1196-210. doi: 10.1016/j.chembiol.2014.07.024. Chem Biol. 2014. PMID: 25237863 Review. - Histone modification enzymes: novel targets for cancer drugs.
Kristeleit R, Stimson L, Workman P, Aherne W. Kristeleit R, et al. Expert Opin Emerg Drugs. 2004 May;9(1):135-54. doi: 10.1517/eoed.9.1.135.32947. Expert Opin Emerg Drugs. 2004. PMID: 15155140 Review. - Small molecule modulators of histone acetylation and methylation: a disease perspective.
Selvi BR, Mohankrishna DV, Ostwal YB, Kundu TK. Selvi BR, et al. Biochim Biophys Acta. 2010 Oct-Dec;1799(10-12):810-28. doi: 10.1016/j.bbagrm.2010.09.005. Epub 2010 Oct 1. Biochim Biophys Acta. 2010. PMID: 20888936 Review. - Erasers of histone acetylation: the histone deacetylase enzymes.
Seto E, Yoshida M. Seto E, et al. Cold Spring Harb Perspect Biol. 2014 Apr 1;6(4):a018713. doi: 10.1101/cshperspect.a018713. Cold Spring Harb Perspect Biol. 2014. PMID: 24691964 Free PMC article. Review. - Screening for compounds that modulate epigenetic regulation of the transcriptome: an overview.
Eglen RM, Reisine T. Eglen RM, et al. J Biomol Screen. 2011 Dec;16(10):1137-52. doi: 10.1177/1087057111417871. Epub 2011 Oct 14. J Biomol Screen. 2011. PMID: 22002420 Review.
Cited by
- Valproic Acid Induces Antimicrobial Compound Production in Doratomyces microspores.
Zutz C, Bacher M, Parich A, Kluger B, Gacek-Matthews A, Schuhmacher R, Wagner M, Rychli K, Strauss J. Zutz C, et al. Front Microbiol. 2016 Apr 13;7:510. doi: 10.3389/fmicb.2016.00510. eCollection 2016. Front Microbiol. 2016. PMID: 27148199 Free PMC article. - Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors.
Culhane JC, Wang D, Yen PM, Cole PA. Culhane JC, et al. J Am Chem Soc. 2010 Mar 10;132(9):3164-76. doi: 10.1021/ja909996p. J Am Chem Soc. 2010. PMID: 20148560 Free PMC article. - Accessing protein methyltransferase and demethylase enzymology using microfluidic capillary electrophoresis.
Wigle TJ, Provencher LM, Norris JL, Jin J, Brown PJ, Frye SV, Janzen WP. Wigle TJ, et al. Chem Biol. 2010 Jul 30;17(7):695-704. doi: 10.1016/j.chembiol.2010.04.014. Chem Biol. 2010. PMID: 20659682 Free PMC article. - Structural Basis of Sirtuin 6-Catalyzed Nucleosome Deacetylation.
Wang ZA, Markert JW, Whedon SD, Yapa Abeywardana M, Lee K, Jiang H, Suarez C, Lin H, Farnung L, Cole PA. Wang ZA, et al. J Am Chem Soc. 2023 Mar 29;145(12):6811-6822. doi: 10.1021/jacs.2c13512. Epub 2023 Mar 17. J Am Chem Soc. 2023. PMID: 36930461 Free PMC article. - Coordinated chromatin control: structural and functional linkage of DNA and histone methylation.
Cheng X, Blumenthal RM. Cheng X, et al. Biochemistry. 2010 Apr 13;49(14):2999-3008. doi: 10.1021/bi100213t. Biochemistry. 2010. PMID: 20210320 Free PMC article. Review.
References
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. - PubMed
- Murray K. The occurrence of ε-N-methyl lysine in histones. Biochemistry. 1964;3:10–15. - PubMed
- Gershey EL, Vidali G, Allfrey VG. Chemical studies of histone acetylation. The occurrence of ε-N-acetyllysine in the f2a1 histone. J. Biol. Chem. 1968;243:5018–5022. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM062437/GM/NIGMS NIH HHS/United States
- R01 GM062437-08/GM/NIGMS NIH HHS/United States
- U54 RR020839/RR/NCRR NIH HHS/United States
- U54 RR020839-056891/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information