BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells - PubMed (original) (raw)
BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells
Matthias D Kaeser et al. J Biol Chem. 2008.
Abstract
The composition of chromatin-remodeling complexes dictates how these enzymes control transcriptional programs and cellular identity. In the present study we investigated the composition of SWI/SNF complexes in embryonic stem cells (ESCs). In contrast to differentiated cells, ESCs have a biased incorporation of certain paralogous SWI/SNF subunits with low levels of BRM, BAF170, and ARID1B. Upon differentiation, the expression of these subunits increases, resulting in a higher diversity of compositionally distinct SWI/SNF enzymes. We also identified BRD7 as a novel component of the Polybromo-associated BRG1-associated factor (PBAF) complex in both ESCs and differentiated cells. Using short hairpin RNA-mediated depletion of BRG1, we showed that SWI/SNF can function as both a repressor and an activator in pluripotent cells, regulating expression of developmental modifiers and signaling components such as Nodal, ADAMTS1, BMI-1, CRABP1, and thyroid releasing hormone. Knockdown studies of PBAF-specific BRD7 and of a signature subunit within the BAF complex, ARID1A, showed that these two subcomplexes affect SWI/SNF target genes differentially, in some cases even antagonistically. This may be due to their different biochemical properties. Finally we examined the role of SWI/SNF in regulating its target genes during differentiation. We found that SWI/SNF affects recruitment of components of the preinitiation complex in a promoter-specific manner to modulate transcription positively or negatively. Taken together, our results provide insight into the function of compositionally diverse SWI/SNF enzymes that underlie their inherent gene-specific mode of action.
Figures
FIGURE 1.
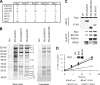
SWI/SNF composition in embryonic stem cells. A, schematic diagram of known SWI/SNF complexes. Signature subunits for BAF complexes are the mutually exclusive ARID1A and ARID1B. BAF complexes can contain either ATPase, BRG1 or BRM. PBAF complexes are characterized by the presence of ARID2 and Polybromo and are restricted to BRG1. B, protein levels of ectopically expressed SWI/SNF subunits. R201 cells were infected with lentiviruses expressing FLAG-tagged proteins as indicated. Western blots were performed on whole cell lysates. C, composition of purified complexes. Using FLAG affinity purification, SWI/SNF was isolated from R201 expressing the indicated subunits, resolved by SDS-PAGE, and silver-stained. Identification of indicated subunits was performed by Western blot and mass spectroscopy. 1, ARID1A; 2, ARID2, Polybromo; 3, BRG1; 4, BAF170; 5, BAF155; 6, BAF60; 7, BAF57; 8, BAF53. D, Western blot analysis of purified complexes. SWI/SNF quantities were adjusted to contain similar BAF60 levels.ctrl, control; Pb, Polybromo.
FIGURE 2.
SWI/SNF compositional changes during RA differentiation. A, complex composition of purified complexes. SWI/SNF was isolated from R201 cells expressing BAF47-FLAG (ESC) and R1 ESCs with BAF47-FLAG (2d RA and 6d RA) induced for differentiation as indicated. Purified complexes were resolved by SDS-PAGE and silver-stained. Asterisks designate nonspecific bands (see also Fig. 1_C_). B, changes in SWI/SNF subunits during differentiation. Complexes described in A were subjected to Western blotting. C, transcriptional changes for SWI/SNF subunits. RNA was isolated during an RA differentiation time course at the indicated time points, and expression levels of indicated subunits were measured by reverse transcription-quantitative PCR and normalized using β-actin. D, schematic representation of compositional changes in SWI/SNF triggered by RA differentiation. 6-day retinoic acid treatment leads to increased expression of BRM, ARID1B, and BAF170 but to a decrease of ARID1A. Pb, Polybromo; ind., induction; d, days. The data presented are the mean values + S.E. from two independent experiments.
FIGURE 3.
Identification of novel SWI/SNF subunits. A, polypeptides consistently found in MudPIT spectroscopy. SWI/SNF was purified as indicated in Fig. 1_C_. Listed are the numbers of individual peptides that could be unequivocally assigned to a particular polypeptide. Differences in peptide recovery between individual samples also reflect variations in total amount of complex subjected to mass spectroscopy. B, putative new subunits co-purify with SWI/SNF. Associated complexes were purified from R218 cells expressing 2FLAG-tagged proteins as indicated, resolved by SDS-PAGE, and stained by silver or Coomassie Blue as indicated. Indicated bands were excised and identified by mass spectroscopy according to highest Mowse score (in parentheses).1, ARID1A (1274); 2, ARID2 (654); Polybromo (1233);3, BRG1 (2131); 4, BAF155 (1751); 5, BRD7 (273);6, BAF60A (797); 7, BAF57 (959); 8, DPF2 (531); BAF53 (353); 9, BAF47 (519); 10, β-actin (719);11, BCL7C (347). Asterisks indicate the tagged subunit of each preparation. C, BRD7 is a PBAF-specific subunit. Lysate and FLAG-immunoprecipitated complexes were resolved by SDS-PAGE and subjected to Western blotting using the indicated antibodies. D, ESC PBAF exhibits different ATP hydrolysis activities than total ESC SWI/SNF. Purified complexes from pluripotent cells were incubated in the presence or absence of DNA. ATPase activity was assessed by calculating the percentage of hydrolyzed32P-labeled phosphate after separation by thin layer chromatography. The inset contains a Western blot for BRG1 of both ATPase reactions. ctrl, control.
FIGURE 4.
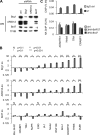
BRD7 regulates a subset of SWI/SNF target genes. A, shRNA-mediated inhibition of specific SWI/SNF subunits. R218 cells were infected with lentiviral shRNA against the indicated subunit or scrambled control. Proteins were extracted and detected by Western blot. B, quantification of SWI/SNF target gene expression. RNA was isolated from cells treated with shRNA viruses and quantified by reverse transcription-quantitative PCR. Target gene expression was normalized to β-actin, and induction over a control virus was plotted on a log scale. Indicated p values were calculated using Student's t test (two independent experiments). C, ChIP assay to quantify SWI/SNF binding to the promoter region of indicated target genes. For the first panel, lysates from cross-linked R218 cells were precipitated using antibodies directed against BRG1 (J1) or a non-expressed epitope (IgG ctrl). For the second panel, ChIP using an antibody directed to the HA epitope was used to precipitate from lysates of cells stably expressing the indicated HA-tagged subunit. Precipitated DNA is expressed as percentage of input DNA. WB, Western blot; ctrl, control; ko, knock-out; IN, input. The data presented are the mean values + S.E. from two independent experiments.
FIGURE 5.
SWI/SNF and differentiation dictate expression by regulating PIC occupancy. A, quantification of target gene expression. RNA was extracted from R218 cells differentiated with RA for the indicated days. mRNA levels were measured by quantitative reverse transcription-PCR and normalized using β-actin. B, ChIP assay to quantify changes in occupancy at the genomic locus of the target genes. Antibodies directed against the indicated proteins/modifications were used to precipitate from lysates of differentiated cross-linked cells. Precipitated DNA is expressed as percentage of input DNA. N, p > 0.15; *, p < 0.15; **, p < 0.05. C, genomic localization of SWI/SNF by ChIP. R218 cells expressing 3HA-BAF57 were differentiated for the indicated time, and ChIP was performed. Primer sets used for amplification were spread over 7 kb as indicated. Precipitated DNA is expressed as percentage of input DNA.D, quantification of target gene expression. RNA was extracted from R218 cells infected with lentiviral shRNAs targeting BRG1 or control (GLUT4) for 96 h. mRNA levels were measured by quantitative reverse transcription-PCR and normalized using β-actin. E, ChIP assay to quantify changes in occupancy at the genomic locus of the target genes Trh and_Adamts1_. Antibodies directed against the indicated proteins/modifications were used to precipitate from lysates of cross-linked cells infected as in A. Occupancy is expressed as -fold change over control shRNA. For p values, see B. Ctrl, control;d, days; ind., induction. The data presented are the mean values + S.E. from two independent experiments.
Similar articles
- Novel Interactions between the Human T-Cell Leukemia Virus Type 1 Antisense Protein HBZ and the SWI/SNF Chromatin Remodeling Family: Implications for Viral Life Cycle.
Alasiri A, Abboud Guerr J, Hall WW, Sheehy N. Alasiri A, et al. J Virol. 2019 Jul 30;93(16):e00412-19. doi: 10.1128/JVI.00412-19. Print 2019 Aug 15. J Virol. 2019. PMID: 31142665 Free PMC article. - Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes.
Alpsoy A, Dykhuizen EC. Alpsoy A, et al. J Biol Chem. 2018 Mar 16;293(11):3892-3903. doi: 10.1074/jbc.RA117.001065. Epub 2018 Jan 26. J Biol Chem. 2018. PMID: 29374058 Free PMC article. - BAF60A mediates interactions between the microphthalmia-associated transcription factor and the BRG1-containing SWI/SNF complex during melanocyte differentiation.
Aras S, Saladi SV, Basuroy T, Marathe HG, Lorès P, de la Serna IL. Aras S, et al. J Cell Physiol. 2019 Jul;234(7):11780-11791. doi: 10.1002/jcp.27840. Epub 2018 Dec 4. J Cell Physiol. 2019. PMID: 30515787 Free PMC article. - BAFfling pathologies: Alterations of BAF complexes in cancer.
Arnaud O, Le Loarer F, Tirode F. Arnaud O, et al. Cancer Lett. 2018 Apr 10;419:266-279. doi: 10.1016/j.canlet.2018.01.046. Epub 2018 Jan 31. Cancer Lett. 2018. PMID: 29374542 Review. - Preclinical evidence in the assembly of mammalian SWI/SNF complexes: Epigenetic insights and clinical perspectives in human lung disease therapy.
Trejo-Villegas OA, Heijink IH, Ávila-Moreno F. Trejo-Villegas OA, et al. Mol Ther. 2024 Aug 7;32(8):2470-2488. doi: 10.1016/j.ymthe.2024.06.026. Epub 2024 Jun 22. Mol Ther. 2024. PMID: 38910326 Review.
Cited by
- Genome-scale study of transcription factor expression in the branching mouse lung.
Herriges JC, Yi L, Hines EA, Harvey JF, Xu G, Gray PA, Ma Q, Sun X. Herriges JC, et al. Dev Dyn. 2012 Sep;241(9):1432-53. doi: 10.1002/dvdy.23823. Epub 2012 Jul 20. Dev Dyn. 2012. PMID: 22711520 Free PMC article. - Oncogene Ras/phosphatidylinositol 3-kinase signaling targets histone H3 acetylation at lysine 56.
Liu Y, Wang DL, Chen S, Zhao L, Sun FL. Liu Y, et al. J Biol Chem. 2012 Nov 30;287(49):41469-80. doi: 10.1074/jbc.M112.367847. Epub 2012 Sep 16. J Biol Chem. 2012. PMID: 22982396 Free PMC article. - BRD7 as key factor in PBAF complex assembly and CD8+ T cell differentiation.
Huang F, Lin Y, Qiao Y, Yuan Y, Zhong Z, Luo B, Wu Y, Liu J, Chen J, Zhang W, Zhang H, Liu B. Huang F, et al. JCI Insight. 2024 Jul 2;9(15):e171605. doi: 10.1172/jci.insight.171605. JCI Insight. 2024. PMID: 38954484 Free PMC article. - Human T-lymphotropic virus type 1 transcription and chromatin-remodeling complexes.
Easley R, Carpio L, Guendel I, Klase Z, Choi S, Kehn-Hall K, Brady JN, Kashanchi F. Easley R, et al. J Virol. 2010 May;84(9):4755-68. doi: 10.1128/JVI.00851-09. Epub 2010 Feb 17. J Virol. 2010. PMID: 20164218 Free PMC article. - A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells.
Gatchalian J, Malik S, Ho J, Lee DS, Kelso TWR, Shokhirev MN, Dixon JR, Hargreaves DC. Gatchalian J, et al. Nat Commun. 2018 Dec 3;9(1):5139. doi: 10.1038/s41467-018-07528-9. Nat Commun. 2018. PMID: 30510198 Free PMC article.
References
- Muller, C., and Leutz, A. (2001) Curr. Opin. Genet. Dev. 11 167–174 - PubMed
- Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., Fry, B., Meissner, A., Wernig, M., Plath, K., Jaenisch, R., Wagschal, A., Feil, R., Schreiber, S. L., and Lander, E. S. (2006) Cell 125 315–326 - PubMed
- Niwa, H. (2007) Development 134 635–646 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous