MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis - PubMed (original) (raw)
MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis
Li-Xu Yan et al. RNA. 2008 Nov.
Abstract
To investigate the global expression profile of miRNAs in primary breast cancer (BC) and normal adjacent tumor tissues (NATs) and its potential relevance to clinicopathological characteristics and patient survival, the genome-wide expression profiling of miRNAs in BC was investigated using a microarray containing 435 mature human miRNA oligonucleotide probes. Nine miRNAs of hsa-miR-21, hsa-miR-365, hsa-miR-181b, hsa-let-7f, hsa-miR-155, hsa-miR-29b, hsa-miR-181d, hsa-miR-98, and hsa-miR-29c were observed to be up-regulated greater than twofold in BC compared with NAT, whereas seven miRNAs of hsa-miR-497, hsa-miR-31, hsa-miR-355, hsa-miR-320, rno-mir-140, hsa-miR-127 and hsa-miR-30a-3p were observed to be down-regulated greater than twofold. The most significantly up-regulated miRNAs, hsa-mir-21 (miR-21), was quantitatively analyzed by TaqMan real-time PCR in 113 BC tumors. Interestingly, among the 113 BC cases, high level expression of miR-21 was significantly correlated with advanced clinical stage (P = 0.006, Fisher's exact text), lymph node metastasis (P = 0.007, Fisher's exact text), and shortened survival of the patients (hazard ratio [HR]=5.476, P < 0.001). Multivariate Cox regression analysis revealed this prognostic impact (HR=4.133, P = 0.001) to be independent of disease stage (HR=2.226, P = 0.013) and histological grade (HR=3.681, P = 0.033). This study could identify the differentiated miRNAs expression profile in BC and reveal that miR-21 overexpression was correlated with specific breast cancer biopathologic features, such as advanced tumor stage, lymph node metastasis, and poor survival of the patients, indicating that miR-21 may serve as a molecular prognostic marker for BC and disease progression.
Figures
FIGURE 1.
Unsupervised hierarchical cluster analysis of 16 differentially expressed miRNAs in BC versus NAT, as was determined by SAM analysis. The heat-map shows that BC segregates from NAT based on a 16-gene miRNA profile. Blue indicates low expression and yellow indicates high expression relative to the median. (Rows) Differentially expressed miRNAs; (columns) tissue samples. 42T, 53T, 54T, 58T, 59T, 60T, 62T, and 63T are 8 BC samples. 1P, 2P, and 3P are normal adjacent tissues. 1P is the NAT of 60T, 2P is a pool of NATs for 42T, 53T, 62T, and 63T, while 3P is pooled NATs from 54T, 58T, and 59T. 42T-1 and 42T-2 represent two replicates of the microarray results for sample 42T, and the rest can be deduced accordingly.
FIGURE 2.
Comparison of miRNA fold changes by miRNA microarray and RT-PCR. The fold change in expression was determined for 60T (BC tissue) in comparison with 1P (corresponding NAT). Microarray fold change was calculated by the ratio of miRNA signal intensities in BRAC versus NAT. RT-PCR fold change is the RPCR value (see Materials and Methods). All of the six comparisons are consistent.
FIGURE 3.
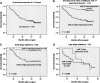
Comparison of miR-21 expression pattern using total RNA extracted from matched samples of FFPETs and frozen tissues. (A) Comparison of Δ_CT_ values of miR-21 assays from paired FFPE and frozen tissue. Δ_CT_ miR-21 = CT miR-21 − CT U6. Identical amount of total RNA was employed in each assay. R 2 is 0.91 between the two kinds of tissues. (B) Comparison of CT values of U6 snRNA assays from paired FFPET and frozen tissue. R 2 is 0.72 between the two kinds of tissues.
FIGURE 4.
miR-21 is differentially expressed between BC and NATs. Data obtained by real-time RT-PCR amplification of miR-21 are plotted. miR-21 is normalized by U6 snRNA. Δ_CT_ = CT miR-21 − CT U6. It is known that the logarithm of the original concentration of the template is the inverse ratio of CT value. In order to demonstrate clearly, we used “−mean ± SE” to describe the expression level of miR-21. The Δ_CT_ of miR-21 in BC (−8.75 ± 0.80, −mean ± SE) was significantly higher than in NATs (−10.04 ± 0.76, −mean ± SE) (P < 0.001, t = −4.84, paired-samples _t_-test). Boxes represent mean. Error bars represent SE.
FIGURE 5.
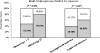
Correlations between miR-21 expression level, clinical stage, and lymph node status of BC patients. In 24 cases with stage III, 19 (79.17%) exhibited high miR-21 expression in BC, whereas in 89 of stages I and II, only 42 (47.19%) presented high level miR-21 expression (P = 0.006, Fisher's Exact Test). In 64 cases of BC with lymph node metastases, 42 (60.87%) presented high miR-21 expression; in contrast, of the 49 cases of BC without lymph node metastasis, only 19 (38.78%) presented high miR-21 expression level (P = 0.007, Fisher's Exact Test).
FIGURE 6.
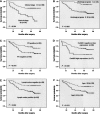
Kaplan–Meier survival curve and log-rank test for BC patients classified as showing either high or low miR-21 expression. (A) Kaplan–Meier curve for 5-yr overall survival rates (64.60%) of 113 patients with BC. (B) High expression of miR-21 (n = 61), group with expression ratio ≥ mean ratio (1.741). mir-21 low expression (n = 52), group with expression ratio < mean ratio. miR-21 expression had a significant (log-rank, P < 0.001) relationship with patient survival. (C) Cases stratified by clinical stage. Within the early stage stratum, miR-21 expression exhibited a significant (P < 0.001; log-rank test) relationship with patient survival. (D) Within the late stage stratum, miR-21 expression did not show a statistical (P = 0.996; log-rank test) relationship with patient survival.
FIGURE 7.
Kaplan–Meier survival curves for BC patients. (A) BC patients were classified as either early or late clinical stage. Clinical stage exhibited a significant (P < 0.001, log-rank) relationship with patient survival. (B) BC patients were classified as histological grade I and II or grade III. Histological grade showed a significant (P = 0.002, log-rank) relationship with patient survival. (C) Kaplan–Meier survival curve for BC patients classified as PR positive or negative. PR expression status was significantly associated with patient survival (log-rank, P = 0.016). (D) Kaplan–Meier survival curve for BC patients classified as showing either high or low CerbB2 expression. CerbB2 status was found to be strongly associated (log-rank, P = 0.04) with patient survival. (E) Kaplan–Meier survival curve for BC patients classified as either lymph node metastasis positive or negative. Lymph node metastasis status was found to be strongly associated (log-rank, P = 0.004) with patient survival. (F) Kaplan–Meier survival curve for BC patients classified as older (≥ median age, 48 yr) or younger (<48 yr). A significant difference in patient survival was observed between the different age groups (log-rank, P = 0.012). (_x_-axis) Survival time after surgery; (_y_-axis) percentage of survivors.
Similar articles
- MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma.
Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF, Huang WZ, Wang WS, Jiang TF, Wu BL, Li EM, Xu LY. Lin RJ, et al. J Surg Oncol. 2012 Feb;105(2):175-82. doi: 10.1002/jso.22066. Epub 2011 Aug 22. J Surg Oncol. 2012. PMID: 21882196 - miRNA expression in breast cancer varies with lymph node metastasis and other clinicopathologic features.
Wang B, Li J, Sun M, Sun L, Zhang X. Wang B, et al. IUBMB Life. 2014 May;66(5):371-7. doi: 10.1002/iub.1273. Epub 2014 May 20. IUBMB Life. 2014. PMID: 24846313 - The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma.
Zhi F, Chen X, Wang S, Xia X, Shi Y, Guan W, Shao N, Qu H, Yang C, Zhang Y, Wang Q, Wang R, Zen K, Zhang CY, Zhang J, Yang Y. Zhi F, et al. Eur J Cancer. 2010 Jun;46(9):1640-9. doi: 10.1016/j.ejca.2010.02.003. Epub 2010 Mar 8. Eur J Cancer. 2010. PMID: 20219352 - Candidate miRNAs in human breast cancer biomarkers: a systematic review.
Adhami M, Haghdoost AA, Sadeghi B, Malekpour Afshar R. Adhami M, et al. Breast Cancer. 2018 Mar;25(2):198-205. doi: 10.1007/s12282-017-0814-8. Epub 2017 Nov 3. Breast Cancer. 2018. PMID: 29101635 Review. - Prognostic value of microRNA expression pattern in upper tract urothelial carcinoma.
Izquierdo L, Ingelmo-Torres M, Mallofré C, Lozano JJ, Verhasselt-Crinquette M, Leroy X, Colin P, Comperat E, Roupret M, Alcaraz A, Mengual L. Izquierdo L, et al. BJU Int. 2014 May;113(5):813-21. doi: 10.1111/bju.12551. BJU Int. 2014. PMID: 24180461 Review.
Cited by
- MicroRNA expression aids the preoperative diagnosis of pancreatic ductal adenocarcinoma.
Panarelli NC, Chen YT, Zhou XK, Kitabayashi N, Yantiss RK. Panarelli NC, et al. Pancreas. 2012 Jul;41(5):685-90. doi: 10.1097/MPA.0b013e318243a905. Pancreas. 2012. PMID: 22466166 Free PMC article. - MicroRNA Changes in Gastric Carcinogenesis: Differential Dysregulation during Helicobacter pylori and EBV Infection.
Prinz C, Mese K, Weber D. Prinz C, et al. Genes (Basel). 2021 Apr 19;12(4):597. doi: 10.3390/genes12040597. Genes (Basel). 2021. PMID: 33921696 Free PMC article. Review. - Oncogenic miR-181a/b affect the DNA damage response in aggressive breast cancer.
Bisso A, Faleschini M, Zampa F, Capaci V, De Santa J, Santarpia L, Piazza S, Cappelletti V, Daidone M, Agami R, Del Sal G. Bisso A, et al. Cell Cycle. 2013 Jun 1;12(11):1679-87. doi: 10.4161/cc.24757. Epub 2013 May 1. Cell Cycle. 2013. PMID: 23656790 Free PMC article. - An image-based biosensor assay strategy to screen for modulators of the microRNA 21 biogenesis pathway.
Shum D, Bhinder B, Radu C, Farazi T, Landthaler M, Tuschl T, Calder P, Ramirez CN, Djaballah H. Shum D, et al. Comb Chem High Throughput Screen. 2012 Aug;15(7):529-41. doi: 10.2174/138620712801619131. Comb Chem High Throughput Screen. 2012. PMID: 22540737 Free PMC article. - The Application of Next-Generation Sequencing to Define Factors Related to Oral Cancer and Discover Novel Biomarkers.
Kim S, Lee JW, Park YS. Kim S, et al. Life (Basel). 2020 Oct 2;10(10):228. doi: 10.3390/life10100228. Life (Basel). 2020. PMID: 33023080 Free PMC article. Review.
References
- Ambros V. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell. 2003;113:673–676. - PubMed
- Bloomston M., Frankel W.L., Petrocca F., Volinia S., Alder H., Hagan J.P., Liu C.G., Bhatt D., Taccioli C., Croce C.M. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. - PubMed
- Bundred N.J. Prognostic and predictive factors in breast cancer. Cancer Treat. Rev. 2001;27:137–142. - PubMed
- Caldas C., Brenton J.D. Sizing up miRNAs as cancer genes. Nat. Med. 2005;11:712–714. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical