A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence - PubMed (original) (raw)
A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence
Joshua J Forman et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Recognition sites for microRNAs (miRNAs) have been reported to be located in the 3' untranslated regions of transcripts. In a computational screen for highly conserved motifs within coding regions, we found an excess of sequences conserved at the nucleotide level within coding regions in the human genome, the highest scoring of which are enriched for miRNA target sequences. To validate our results, we experimentally demonstrated that the let-7 miRNA directly targets the miRNA-processing enzyme Dicer within its coding sequence, thus establishing a mechanism for a miRNA/Dicer autoregulatory negative feedback loop. We also found computational evidence to suggest that miRNA target sites in coding regions and 3' UTRs may differ in mechanism. This work demonstrates that miRNAs can directly target transcripts within their coding region in animals, and it suggests that a complete search for the regulatory targets of miRNAs should be expanded to include genes with recognition sites within their coding regions. As more genomes are sequenced, the methodological approach that we used for identifying motifs with high sequence conservation will be increasingly valuable for detecting functional sequence motifs within coding regions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Motifs with a high SLCS are enriched for miRNA target sequences. The distribution of non-zero SLCSs (black bars) from a 17-genome alignment and the distribution that resulted from an analysis of a randomly permuted genome are plotted. The distribution demonstrates that there are significantly more high-scoring motifs within coding regions compared with the distribution of scores from a randomized alignment (white bars). The highest-scoring sequence motifs (Inset) are enriched for miRNA target sequences. Results are filtered so that conserved motifs are removed if they are within 3 bp of a higher-scoring motif.
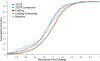
Fig. 2.
Highly conserved let-7 targets within coding regions are functional. Cultured human fibroblasts were transfected with pre-let-7b or a control RNA. Twenty-four hours after transfection, RNA was isolated, labeled, and hybridized to a whole-genome microarray. The fold change in expression between cells transfected with let-7b or the control was determined. The cumulative distribution of the fraction of genes down-regulated in the microarray data shows a significant difference between a random set of genes and genes with target sites in their 3′ UTR (P = 0.014, one-tailed Kolmogorov–Smirnov test), but not with genes with target sites in their coding region (P = 0.26). Genes with highly conserved target sites in their coding region, however, are significantly different from the set of random genes (P < 10−4), as are genes with highly conserved target sites in their 3′ UTRs (P < 10−3).
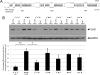
Fig. 3.
The microRNA let-7 targets the coding region of Dicer. (A) Dicer contains three highly conserved let-7 target sites. Alignments between the let-7 miRNA and the target sites are shown. The first two sites are conserved in 16 of 17 genomes, and the last is conserved in all 17 genomes. The coding sequence of Dicer is shown above let-7b. (B) HEK 293 cells were cotransfected with Dicer and with let-7b pre-miRNA or a pre-miRNA control. Several variants of the Dicer construct were used, from which various combinations of the let-7 sites had been abrogated by site-directed mutagenesis. (Upper) An immunoblot of Dicer levels and GAPDH levels. Wild-type Dicer is in lanes 1 and 2, fully mutated Dicer is in lanes 3 and 4, and each subsequent pair of lanes is marked to indicate which sites were intact (✓) and which were abrogated (×). Cellular lysates were analyzed by immunoblotting with antibodies to the FLAG epitope (upper band) or GAPDH (lower band). (Lower) The fold reduction in the presence versus absence of exogenous let-7 for each Dicer variant. Means (n = 4) and standard errors are plotted. Wild-type Dicer is significantly down-regulated by _let_-7 compared with the Dicer variant with all three target sites abrogated (two-tailed t test, P < 0.01). The Dicer variant with the first two target sites intact is also significantly down-regulated (P = 0.02). The Dicer variant with the first and third target sites intact is down-regulated with the next-highest magnitude, but the effect did not reach significance. Dicer variants with a single site intact are down-regulated less strongly than variants with two sites.
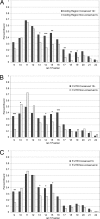
Fig. 4.
Recognition sites at various positions within a transcript have different patterns of miRNA binding. Sites complementary to the let-7 seed were extracted from 3′ UTRs (A), coding regions (B), and 5′ UTRs (C), along with additional upstream sequence, and computationally folded to a consensus let-7 miRNA (see Materials and Methods). The fraction of target–seed interactions in which a particular base pair was bound was determined for each position of the miRNA outside of the seed. The fraction bound was compared for highly conserved versus poorly conserved target seeds. Numbers indicate the number of genomes in which a seed was conserved; 12+ indicates indicates conservation in 12 or more genomes, 3− indicates conservation in three or fewer genomes. Statistical comparisons were performed by using the Fisher exact test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Different cutoffs for the number of genomes in the “conserved” and “nonconserved” groups were used for each search region, and target seeds of 6 bp were used for 5′ UTRs to ensure an adequate number of sequences on which to perform statistical analysis (
Fig. S3
shows results for various conservation levels).
Similar articles
- let-7 regulates Dicer expression and constitutes a negative feedback loop.
Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. Tokumaru S, et al. Carcinogenesis. 2008 Nov;29(11):2073-7. doi: 10.1093/carcin/bgn187. Epub 2008 Aug 11. Carcinogenesis. 2008. PMID: 18700235 - Bioinformatic discovery of microRNA precursors from human ESTs and introns.
Li SC, Pan CY, Lin WC. Li SC, et al. BMC Genomics. 2006 Jul 3;7:164. doi: 10.1186/1471-2164-7-164. BMC Genomics. 2006. PMID: 16813663 Free PMC article. - Computational identification of Drosophila microRNA genes.
Lai EC, Tomancak P, Williams RW, Rubin GM. Lai EC, et al. Genome Biol. 2003;4(7):R42. doi: 10.1186/gb-2003-4-7-r42. Epub 2003 Jun 30. Genome Biol. 2003. PMID: 12844358 Free PMC article. - miRNA_Targets: a database for miRNA target predictions in coding and non-coding regions of mRNAs.
Kumar A, Wong AK, Tizard ML, Moore RJ, Lefèvre C. Kumar A, et al. Genomics. 2012 Dec;100(6):352-6. doi: 10.1016/j.ygeno.2012.08.006. Epub 2012 Aug 25. Genomics. 2012. PMID: 22940442 - MicroRNAs tune cerebral cortical neurogenesis.
Volvert ML, Rogister F, Moonen G, Malgrange B, Nguyen L. Volvert ML, et al. Cell Death Differ. 2012 Oct;19(10):1573-81. doi: 10.1038/cdd.2012.96. Epub 2012 Aug 3. Cell Death Differ. 2012. PMID: 22858543 Free PMC article. Review.
Cited by
- Genome-wide and cell-type-selective profiling of in vivo small noncoding RNA:target RNA interactions by chimeric RNA sequencing.
Li X, Mills WT 4th, Jin DS, Meffert MK. Li X, et al. Cell Rep Methods. 2024 Aug 19;4(8):100836. doi: 10.1016/j.crmeth.2024.100836. Epub 2024 Aug 9. Cell Rep Methods. 2024. PMID: 39127045 Free PMC article. - MicroRNAs as Biomarkers and Therapeutic Targets in Inflammation- and Ischemia-Reperfusion-Related Acute Renal Injury.
Wu YL, Li HF, Chen HH, Lin H. Wu YL, et al. Int J Mol Sci. 2020 Sep 14;21(18):6738. doi: 10.3390/ijms21186738. Int J Mol Sci. 2020. PMID: 32937906 Free PMC article. Review. - The potential role of microRNAs in regulating gonadal sex differentiation in the chicken embryo.
Cutting AD, Bannister SC, Doran TJ, Sinclair AH, Tizard MV, Smith CA. Cutting AD, et al. Chromosome Res. 2012 Jan;20(1):201-13. doi: 10.1007/s10577-011-9263-y. Chromosome Res. 2012. PMID: 22161018 - Insect microRNAs: biogenesis, expression profiling and biological functions.
Lucas K, Raikhel AS. Lucas K, et al. Insect Biochem Mol Biol. 2013 Jan;43(1):24-38. doi: 10.1016/j.ibmb.2012.10.009. Epub 2012 Nov 16. Insect Biochem Mol Biol. 2013. PMID: 23165178 Free PMC article. Review. - Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs.
Londin E, Loher P, Telonis AG, Quann K, Clark P, Jing Y, Hatzimichael E, Kirino Y, Honda S, Lally M, Ramratnam B, Comstock CE, Knudsen KE, Gomella L, Spaeth GL, Hark L, Katz LJ, Witkiewicz A, Rostami A, Jimenez SA, Hollingsworth MA, Yeh JJ, Shaw CA, McKenzie SE, Bray P, Nelson PT, Zupo S, Van Roosbroeck K, Keating MJ, Calin GA, Yeo C, Jimbo M, Cozzitorto J, Brody JR, Delgrosso K, Mattick JS, Fortina P, Rigoutsos I. Londin E, et al. Proc Natl Acad Sci U S A. 2015 Mar 10;112(10):E1106-15. doi: 10.1073/pnas.1420955112. Epub 2015 Feb 23. Proc Natl Acad Sci U S A. 2015. PMID: 25713380 Free PMC article.
References
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. - PubMed
- Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. - PubMed
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5T32 CA009528/CA/NCI NIH HHS/United States
- 1R01 GM081686/GM/NIGMS NIH HHS/United States
- R01 GM086465/GM/NIGMS NIH HHS/United States
- P50 GM071508/GM/NIGMS NIH HHS/United States
- T32 CA009528/CA/NCI NIH HHS/United States
- R01 GM081686/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials