Frequency-modulated nuclear localization bursts coordinate gene regulation - PubMed (original) (raw)
Frequency-modulated nuclear localization bursts coordinate gene regulation
Long Cai et al. Nature. 2008.
Abstract
In yeast, the transcription factor Crz1 is dephosphorylated and translocates into the nucleus in response to extracellular calcium. Here we show, using time-lapse microscopy, that Crz1 exhibits short bursts of nuclear localization (typically lasting 2 min) that occur stochastically in individual cells and propagate to the expression of downstream genes. Strikingly, calcium concentration controls the frequency, but not the duration, of localization bursts. Using an analytic model, we also show that this frequency modulation of bursts ensures proportional expression of multiple target genes across a wide dynamic range of expression levels, independent of promoter characteristics. We experimentally confirm this theory with natural and synthetic Crz1 target promoters. Another stress-response transcription factor, Msn2, exhibits similar, but largely uncorrelated, localization bursts under calcium stress suggesting that frequency-modulation regulation of localization bursts may be a general control strategy used by the cell to coordinate multi-gene responses to external signals.
Figures
Figure 1
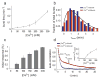
Crz1 undergoes bursts of nuclear localization in response to calcium. a, In the presence of extracellular Ca2+, Crz1 is dephosphorylated and translocates into the nucleus. b, Filmstrip showing yeast cells with Crz1-GFP before and after addition of 200mM extracellular calcium (yellow square). Frames displayed here are separated by 4.5 min, but actual time resolution is higher. c,d Two single cell time traces showing Crz1 localization behavior of the two cells in b. Note that there is a synchronized initial burst of nuclear localization followed by subsequent unsynchronized isolated and cluster bursts of localization. Individual burst duration, τburst, and cluster duration, τcluster, as well as the delay between calcium addition and the initial response, τdelay, are defined on the traces. e, Averaged localization trace shows how single-cell burst dynamics yield partial adaptation across a population of cells.
Figure 2
Calcium modulates the frequency, but not the duration, of Crz1 nuclear localization bursts. a, Frequency of bursts increases with calcium concentration (error bars calculated by using different thresholds for burst determination, see Supplementary Information). b, Burst duration is independent of calcium concentration. Normalized histograms, h(t), of total burst duration at two calcium concentrations are both well-fit by h(t) = t e_−_t/τ, with τ = 70 sec (black line). c. The proportion of cells that respond rapidly to extracellular calcium increases with the calcium concentration. Bars represent the fraction of cells with nuclear-localized Crz1 within 15 minutes of addition of calcium. d, Average auto-correlation functions of localization trajectories (N=58 and 85 cells respectively) from a population of cells at two calcium concentrations. At low Ca2+ concentrations (blue, 50mM), the autocorrelation can be well-fit by a single exponential with timescale τ_burst_≈60s, whereas at high Ca2+ concentrations (red, 200mM), two time scales of fluctuations emerge, τ_burst_≈ 60s and τ_cluster≈_ 720s, corresponding to isolated and clustered bursts, respectively. Inset shows the relative weight of the clustered bursts, which appear at Ca2+ concentrations greater than 100mM and increase in frequency as calcium increases. Error bars are estimated from bootstrap.
Figure 3
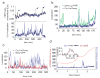
Crz1 localization bursts are partially independent of other cellular processes and affect downstream gene expression. a, Time traces of Crz1-mCherry localization (blue) in arbitrary units and FRET ratio changes (black), indicating intracellular calcium levels. Arrows indicate spontaneous calcium spikes coincident with Crz1 localization bursts. b, Single-cell time traces of Crz1-mCherry (blue) and the Crz1high-GFP mutant (green) with increased affinity to calcineurin. Both Crz1 proteins are expressed and measured simultaneously in the same cell. c, single-cell traces of Crz1-mCherry and Msn2-GFP in the same cell. Note that the two proteins exhibit statistically similar burst-like behavior but only weak correlation. d, Crz1-mCherry localization (blue) increases expression of the Crz1 target synthetic promoter (2X CDRE) (red). Transcriptional bursts in p2XCDRE-venus are preceded by corresponding Crz1 localization bursts. Inset shows positive cross-correlation (_n_=9 cells) between the promoter activity and Crz1 localization with a delay corresponding to target protein maturation. Error bars are estimated from bootstrap.
Figure 4
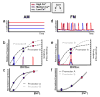
Frequency- versus amplitude –modulation regulation of two hypothetical target genes, labeled A and B (schematic). a, In the amplitude-modulation regulation system, the fraction of nuclear Crz1 (Crz1nuc) changes with calcium, but remains constant over time. b, As such, the histogram of Crz1nuc yields single peaks at calcium-dependent positions. Target gene expression level is proportional to the input functions at these peak positions. c, Because their input functions differ, the normalized rates of A and B expression vary differently with nuclear Crz1, and hence with calcium, yielding different (uncoordinated) expression profiles as a function of calcium. d, In frequency modulation, where Crz1 molecules collectively move between nuclear and cytoplasmic compartments, Crz1nuc is either high or low, during or between bursts, respectively. This graph depicts the limiting case of rapid and complete transitions between two states, but results do not depend on this assumption (see Supplementary Information). e, This yields a bimodal histogram in which the height, but not the position, of the peak is calcium-dependent. f, Consequently, the expression levels A and B are each proportional to burst frequency, and hence to each other, yielding coordinated expression, as shown.
Figure 5
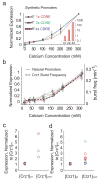
Frequency-modulated bursts coordinate gene expression. a. Measured expression levels of synthetic Crz1 target promoters containing one, two and four CDREs. In each case, expression levels are normalized by their maximum. Note similarity of curves to each other and to burst frequency. Bottom inset shows expression at 200mM calcium normalized by expression of the 1× CDRE. b, Expression profiles of natural target promoters (solid lines) exhibit similar inductions as the Crz1 burst frequency (dashed line). Only ten are plotted here for clarity. Error bars indicate standard error from repeated experiments. Induction curves for 64 genes are shown in Supplementary Fig. 14. c, Expression level of synthetic target genes at wild-type versus overexpressed levels of Crz1. Each promoter is normalized by the maximum expression in wild-type Crz1 levels. Thus the normalized expression levels of one, two and four CDREs at [Crz1]wt each equal 1. If all promoter input functions were identical, then their normalized expression should increase by the same factor when Crz1 is overexpressed. However, the data show a range of fold changes in expression levels, excluding identical input function shapes as an explanation for proportional regulation. d. Same as c. for the natural promoters in b.
Similar articles
- A Noisy Analog-to-Digital Converter Connects Cytosolic Calcium Bursts to Transcription Factor Nuclear Localization Pulses in Yeast.
Hsu IS, Strome B, Plotnikov S, Moses AM. Hsu IS, et al. G3 (Bethesda). 2019 Feb 7;9(2):561-570. doi: 10.1534/g3.118.200841. G3 (Bethesda). 2019. PMID: 30573469 Free PMC article. - Bursting into the nucleus.
Crabtree GR, Graef IA. Crabtree GR, et al. Sci Signal. 2008 Dec 23;1(51):pe54. doi: 10.1126/scisignal.151pe54. Sci Signal. 2008. PMID: 19109237 Free PMC article. Review. - The yeast transcription factor Crz1 is activated by light in a Ca2+/calcineurin-dependent and PKA-independent manner.
Bodvard K, Jörhov A, Blomberg A, Molin M, Käll M. Bodvard K, et al. PLoS One. 2013;8(1):e53404. doi: 10.1371/journal.pone.0053404. Epub 2013 Jan 15. PLoS One. 2013. PMID: 23335962 Free PMC article. - Nuclear localization domains of GATA activator Gln3 are required for transcription of target genes through dephosphorylation in Saccharomyces cerevisiae.
Numamoto M, Tagami S, Ueda Y, Imabeppu Y, Sasano Y, Sugiyama M, Maekawa H, Harashima S. Numamoto M, et al. J Biosci Bioeng. 2015 Aug;120(2):121-7. doi: 10.1016/j.jbiosc.2014.12.017. Epub 2015 Jan 29. J Biosci Bioeng. 2015. PMID: 25641578 - Oscillatory behavior of the nuclear localization of the transcription factors Msn2 and Msn4 in response to stress in yeast.
Jacquet M, Renault G, Lallet S, De Mey J, Goldbeter A. Jacquet M, et al. ScientificWorldJournal. 2003 Jul 1;3:609-12. doi: 10.1100/tsw.2003.47. ScientificWorldJournal. 2003. PMID: 12858011 Free PMC article. Review. No abstract available.
Cited by
- How to regulate a gene: to repress or to activate?
Slavov N, van Oudenaarden A. Slavov N, et al. Mol Cell. 2012 Jun 8;46(5):551-2. doi: 10.1016/j.molcel.2012.05.035. Mol Cell. 2012. PMID: 22681881 Free PMC article. - Pathways to cellular supremacy in biocomputing.
Grozinger L, Amos M, Gorochowski TE, Carbonell P, Oyarzún DA, Stoof R, Fellermann H, Zuliani P, Tas H, Goñi-Moreno A. Grozinger L, et al. Nat Commun. 2019 Nov 20;10(1):5250. doi: 10.1038/s41467-019-13232-z. Nat Commun. 2019. PMID: 31748511 Free PMC article. Review. - Localization of Noise in Biochemical Networks.
Fajiculay E, Hsu CP. Fajiculay E, et al. ACS Omega. 2023 Jan 11;8(3):3043-3056. doi: 10.1021/acsomega.2c06113. eCollection 2023 Jan 24. ACS Omega. 2023. PMID: 36713703 Free PMC article. - Nucleocytoplasmic shuttling of a GATA transcription factor functions as a development timer.
Cai H, Katoh-Kurasawa M, Muramoto T, Santhanam B, Long Y, Li L, Ueda M, Iglesias PA, Shaulsky G, Devreotes PN. Cai H, et al. Science. 2014 Mar 21;343(6177):1249531. doi: 10.1126/science.1249531. Science. 2014. PMID: 24653039 Free PMC article. - A dynamical model reveals gene co-localizations in nucleus.
Kang J, Xu B, Yao Y, Lin W, Hennessy C, Fraser P, Feng J. Kang J, et al. PLoS Comput Biol. 2011 Jul;7(7):e1002094. doi: 10.1371/journal.pcbi.1002094. Epub 2011 Jul 7. PLoS Comput Biol. 2011. PMID: 21760760 Free PMC article.
References
- Cyert MS. Regulation of nuclear localization during signaling. J Biol Chem. 2001;276(24):20805. - PubMed
- Estruch F. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev. 2000;24(4):469. - PubMed
- Kyriakis JM. The integration of signaling by multiprotein complexes containing Raf kinases. Biochim Biophys Acta. 2007;1773(8):1238. - PubMed
- Yoshimoto H, et al. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J Biol Chem. 2002;277(34):31079. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 GM068763/GM/NIGMS NIH HHS/United States
- P50 GM068763-050005/GM/NIGMS NIH HHS/United States
- R01 GM079771-02/GM/NIGMS NIH HHS/United States
- R01GM079771/GM/NIGMS NIH HHS/United States
- R01 GM079771/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases