Intronic Alus influence alternative splicing - PubMed (original) (raw)
Intronic Alus influence alternative splicing
Galit Lev-Maor et al. PLoS Genet. 2008.
Abstract
Examination of the human transcriptome reveals higher levels of RNA editing than in any other organism tested to date. This is indicative of extensive double-stranded RNA (dsRNA) formation within the human transcriptome. Most of the editing sites are located in the primate-specific retrotransposed element called Alu. A large fraction of Alus are found in intronic sequences, implying extensive Alu-Alu dsRNA formation in mRNA precursors. Yet, the effect of these intronic Alus on splicing of the flanking exons is largely unknown. Here, we show that more Alus flank alternatively spliced exons than constitutively spliced ones; this is especially notable for those exons that have changed their mode of splicing from constitutive to alternative during human evolution. This implies that Alu insertions may change the mode of splicing of the flanking exons. Indeed, we demonstrate experimentally that two Alu elements that were inserted into an intron in opposite orientation undergo base-pairing, as evident by RNA editing, and affect the splicing patterns of a downstream exon, shifting it from constitutive to alternative. Our results indicate the importance of intronic Alus in influencing the splicing of flanking exons, further emphasizing the role of Alus in shaping of the human transcriptome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Bioinformatic analysis of Alu insertion within the flanking introns.
Conserved constitutively spliced exons were analyzed for the differences in the location of antisense and sense _Alu_s within the upstream and downstream introns (left and right panels, respectively). The x-axis is the distance in base pairs from the exon; the y-axis is the number of _Alu_s found within this distance. Antisense _Alu_s are marked in blue and sense _Alu_s are marked in red.
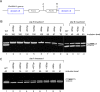
Figure 2. The effect of intronic _Alu_s on the splicing of a flanking exon.
(A) Schematic illustration of the RABL5 minigene containing three exons and two introns. The intronic _Alu_s (1 through 5) are marked by boxes with a point indicating the orientation of the Alu relative to the pre-mRNA. _Alu_4 is an _Alu_Sx inserted between the two arms of _Alu_Jo. (B) The indicated wild-type (wt) and mutant plasmids were transfected into 293T cells, total RNA was extracted, and splicing products were separated on a 2% agarose gel after RT-PCR analysis. Lane 1, splicing products of wt RABL5 minigene. Lanes 2–26, splicing products of the indicated mutants. The following abbreviations were used: Δ indicates deletion of the specified Alu element, X 1w3 indicates replacement of _Alu_1 with _Alu_3 (i.e., the sequence of _Alu_3 was inserted instead of that of _Alu_1 and in the same orientation as _Alu_3), and 1int specifies replacement of the _Alu_1 sequence with a non-Alu intronic fragment. The two mRNA isoforms are shown on the right. Numbers on top of the gel indicate percentage of exon inclusion as determined using ImageJ software. PCR products were sequenced.
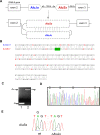
Figure 3. Editing sites within the intronic _Alu_s.
(A) Schematic illustration of exons 2 to 3 of the RABL5 gene. Exons are depicted as black boxes; the intronic Alus, derived from _Alu_Jo and _Alu_Sx, in sense and antisense orientations, respectively, are shown in the middle gray-shaped boxes. The intronic, antisense Alu sequence (_Alu_Sx) is 102 nucleotides downstream of the sense _Alu_Jo and _Alu_Sx is 24 nucleotides upstream of the junction of exon 3. Sense and antisense _Alu_s are expected to form a double-stranded secondary structure, thus allowing RNA editing. (B) Editing sites were inferred from alignment of five cDNAs (accession numbers BC050531, BC038668, BI547904, BI548328, and DB495755) to the human genomic DNA. RNA editing occurs at eight positions within the antisense _Alu_Sx and at eleven positions within the sense _Alu_Jo. Based on these editing sites, the pairing between the sense and antisense Alu sequences was inferred (upper and lower lines, respectively). The region in which editing occurs starts at the middle of the right arm (position 232 in the AluSx consensus) and ends at the beginning of the left arm of _Alu_2 (position 101 in the AluSx consensus). Panel B shows only this corresponding region, while the entire _Alu_-Alu potential dsRNA is shown in Figure S3. (C) To further confirm the editing activity, total RNA was extracted from a neuroblastoma (SH-SY5Y) cell-line and treated with DNaseI, followed by RT-PCR analysis using primers to exon 2 and exon 3, and to intron 2 and exon 3 (lanes 1 and 2, respectively; see also Materials and Methods). The PCR products were sequenced. (D) The upper PCR product shown in panel C lane 2 was cloned and sequenced. The Chromas sequence is shown with the editing sites, found in _Alu_Sx, marked by boxes. (E) Editing in _Alu_2 requires the presence of _Alu_1. Wild type RABL5 minigene (WT) and a mutant in which _Alu_1 was deleted (Δ_Alu_Jo) were transfected into 293T cells. RNA was extracted, treated with DNase I, and amplified using set of primers flanking _Alu_2 and designed to amplify only exogenic transcripts. Sequencing chromatograms of four nucleotides in AluSx are shown (this editing site is also marked in green in panel B).
Figure 4. Distance effect of Alu elements on the alternative splicing pattern.
(A) A schematic illustration of the genomic region between exons 2 and 3 of RABL5 gene. Arrows marked A and B indicate two positions where an intronic sequence was inserted. (B) An 800-nucleotide intronic sequence was inserted in site B. The 800-nucleotide sequence was gradually shortened to the size shown above each lane. The indicated wt and chimeric plasmids were transfected into human 293T cells, total RNA was collected and examined by RT-PCR analysis (lanes 1–9). Lanes 10–12 show insertions of a different sequence, containing 25 nucleotides without any known splicing regulatory sequences, into the same site. This sequence was duplicated and triplicated to generate 50- and 75-nucleotide inserts. These mutant RABL5 minigenes were examined as above. (C) Similar analysis as in panel B, except that the 800-nucleotide sequence and the shorter sequences were inserted into site A. Spliced products are shown on the right and each PCR product was confirmed by sequencing. Splicing products were separated on a 2% agarose gel. Numbers on top of the gel indicate percentage of exon inclusion as determined using ImageJ software.
Figure 5. The effect of _Alu_2 on the alternative exon.
(A) The sequence of the antisense _Alu_2. Mutated putative 5′ss is shown in red. A sequence of 24 nucleotides that was deleted is underlined. Three putative 3′ss that were mutated are in bold and underlined. In yellow are the right and left PPT regions with the downstream AG that were deleted. The green sequence is a stem and loop region of 18 nucleotides that was examined as shown in panel C (referred as ‘A’ region). Underlined in that region are two overlapping SC35 potential binding sites (the gray in the middle indicates the overlap region). (B) The indicated wt and mutant plasmids were transfected into 293T cells, total RNA was extracted, and splicing products were separated on a 2% agarose gel after RT-PCR analysis. Lane 1, splicing products of wt RABL5 minigene. Lanes 2–13, splicing products of the indicated mutants. The two mRNA isoforms are shown on the right. Numbers on top of the gel indicate percentage of exon inclusion, as determined using ImageJ software. (C) The upper part illustrates the putative secondary structure formed by _Alu_Jo and _Alu_Sx, as predicted using the Vienna secondary structure web site (
http://www.tbi.univie.ac.at/RNA
). The green arrows in the right panel indicate the start and end positions of the stem and loop structure, marked as ‘A’. The lower part shows the effect of the wt and mutant plasmids that were analyzed, as in panel B. Lane 1, wilt type. Lane 2, elimination of the two SC35 putative binding sites by mutating their overlapping sequence (AA were mutated to CC). Lane 3, deletion of the entire stem-loop ‘A’ sequence. Lane 4, replacement of the ‘A’ sequence by a random sequence lacking putative splicing binding sites. Lane 5, replacement of the loop sequence by a perfect complementary sequence, so that the ‘A’ element is entirely in a stem structure. Lane 6, disruption of the stem part of ‘A’, so that ‘A’ is entirely in a loop structure.
Similar articles
- Alternative splicing of Alu exons--two arms are better than one.
Gal-Mark N, Schwartz S, Ast G. Gal-Mark N, et al. Nucleic Acids Res. 2008 Apr;36(6):2012-23. doi: 10.1093/nar/gkn024. Epub 2008 Feb 14. Nucleic Acids Res. 2008. PMID: 18276646 Free PMC article. - RNA-editing-mediated exon evolution.
Lev-Maor G, Sorek R, Levanon EY, Paz N, Eisenberg E, Ast G. Lev-Maor G, et al. Genome Biol. 2007;8(2):R29. doi: 10.1186/gb-2007-8-2-r29. Genome Biol. 2007. PMID: 17326827 Free PMC article. - The birth of an alternatively spliced exon: 3' splice-site selection in Alu exons.
Lev-Maor G, Sorek R, Shomron N, Ast G. Lev-Maor G, et al. Science. 2003 May 23;300(5623):1288-91. doi: 10.1126/science.1082588. Science. 2003. PMID: 12764196 - Exonization of transposed elements: A challenge and opportunity for evolution.
Schmitz J, Brosius J. Schmitz J, et al. Biochimie. 2011 Nov;93(11):1928-34. doi: 10.1016/j.biochi.2011.07.014. Epub 2011 Jul 26. Biochimie. 2011. PMID: 21787833 Review. - The origins and implications of Aluternative splicing.
Kreahling J, Graveley BR. Kreahling J, et al. Trends Genet. 2004 Jan;20(1):1-4. doi: 10.1016/j.tig.2003.11.001. Trends Genet. 2004. PMID: 14698612 Review.
Cited by
- Alu-element insertion in an OPA1 intron sequence associated with autosomal dominant optic atrophy.
Gallus GN, Cardaioli E, Rufa A, Da Pozzo P, Bianchi S, D'Eramo C, Collura M, Tumino M, Pavone L, Federico A. Gallus GN, et al. Mol Vis. 2010 Feb 10;16:178-83. Mol Vis. 2010. PMID: 20157369 Free PMC article. - Circular RNA repertoires are associated with evolutionarily young transposable elements.
Gruhl F, Janich P, Kaessmann H, Gatfield D. Gruhl F, et al. Elife. 2021 Sep 20;10:e67991. doi: 10.7554/eLife.67991. Elife. 2021. PMID: 34542406 Free PMC article. - The identification of switch-like alternative splicing exons among multiple samples with RNA-Seq data.
Qin Z, Zhang X. Qin Z, et al. PLoS One. 2017 May 25;12(5):e0178320. doi: 10.1371/journal.pone.0178320. eCollection 2017. PLoS One. 2017. PMID: 28542625 Free PMC article. - Genomic gems: SINE RNAs regulate mRNA production.
Ponicsan SL, Kugel JF, Goodrich JA. Ponicsan SL, et al. Curr Opin Genet Dev. 2010 Apr;20(2):149-55. doi: 10.1016/j.gde.2010.01.004. Epub 2010 Feb 20. Curr Opin Genet Dev. 2010. PMID: 20176473 Free PMC article. Review. - Interaction of hnRNPA1/A2 and DAZAP1 with an Alu-derived intronic splicing enhancer regulates ATM aberrant splicing.
Pastor T, Pagani F. Pastor T, et al. PLoS One. 2011;6(8):e23349. doi: 10.1371/journal.pone.0023349. Epub 2011 Aug 8. PLoS One. 2011. PMID: 21858080 Free PMC article.
References
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. - PubMed
- Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5:773–782. - PubMed
- Sugnet CW, Kent WJ, Ares M, Jr., Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac Symp Biocomput. 2004:66–77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources