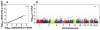Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease - PubMed (original) (raw)
. 2008 Dec;40(12):1461-5.
doi: 10.1038/ng.257. Epub 2008 Sep 25.
Affiliations
- PMID: 18820647
- PMCID: PMC2597056
- DOI: 10.1038/ng.257
Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease
Stefano Romeo et al. Nat Genet. 2008 Dec.
Abstract
Nonalcoholic fatty liver disease (NAFLD) is a burgeoning health problem of unknown etiology that varies in prevalence among ancestry groups. To identify genetic variants contributing to differences in hepatic fat content, we carried out a genome-wide association scan of nonsynonymous sequence variations (n = 9,229) in a population comprising Hispanic, African American and European American individuals. An allele in PNPLA3 (rs738409[G], encoding I148M) was strongly associated with increased hepatic fat levels (P = 5.9 x 10(-10)) and with hepatic inflammation (P = 3.7 x 10(-4)). The allele was most common in Hispanics, the group most susceptible to NAFLD; hepatic fat content was more than twofold higher in PNPLA3 rs738409[G] homozygotes than in noncarriers. Resequencing revealed another allele of PNPLA3 (rs6006460[T], encoding S453I) that was associated with lower hepatic fat content in African Americans, the group at lowest risk of NAFLD. Thus, variation in PNPLA3 contributes to ancestry-related and inter-individual differences in hepatic fat content and susceptibility to NAFLD.
Figures
Figure 1
Whole-genome scan of liver triglyceride content measured by proton magnetic resonance imaging in the Dallas Heart Study (n=2,111). (a) Quantile-quantile plot of P-values. (b) Scatter plot of P-values. The dashed line denotes the Bonferroni corrected significance threshold (P=5.4×10−6).
Figure 2
Association between a sequence variant in PNPLA3 (I148M) and hepatic triglyceride (TG) content. (a) PNPLA3 is a 481 residue protein that contains a patatin-like domain at the N terminus (residues 10-179, UniProt
http://pir.uniprot.org/uniprot/
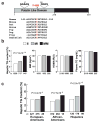
) with consensus sequences for a Ser-Asp catalytic dyad (Gly-X-Ser-X-Gly and Asp-X-Gly/Ala). The I148M substitution (rs738409) is located between the consensus sequences for the catalytic dyad and is highly conserved. (b) Median hepatic TG content, body mass index (BMI), homeostatic model assessment of insulin resistance (HOMA-IR) and plasma TG levels in the Dallas Heart Study. Values for hepatic fat content were compared using ANOVA. Age, gender, BMI, diabetes status, ethanol use and global ancestry were included as covariates in the model. (c) Median hepatic fat contents and _PNPLA3_-I148M genotypes in European-Americans, African-Americans and Hispanics in the Dallas Heart Study. Associations between hepatic fat content and _PNPLA3_-I148M genotypes were tested using ANOVA with age, local ancestry, gender, diabetes status, ethanol intake and BMI as covariates.
Figure 3
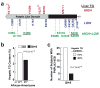
Identification of a PNPLA3 allele (S453I) associated with lower hepatic fat content in African-Americans in the Dallas Heart Study. (a) Exons and flanking introns of PNPLA3 were sequenced in the 32 European-American and African-American men and women and in the 16 Hispanic men and women with the lowest and highest hepatic triglyceride content determined using proton magnetic resonance imaging. The NS variations identified in individuals in only the high, only the low and in both the high and low groups are shown. All the variants not found in both groups were present in only a single subject unless otherwise indicated. The rs numbers for polymorphisms and the oligonucleotides used for PCR-sequencing of the coding regions and are provided in Supplementary Tables 3 online and Supplementary Table 4 online. (b) Median hepatic TG content in African-Americans carrying the wild-type or _PNPLA3_-453I allele. (c) Number of individuals with _PNPLA3_-453I in the upper and lower deciles of hepatic fat content.
Comment in
- The genes that underlie fatty liver disease: the harvest has begun.
Weiskirchen R, Wasmuth HE. Weiskirchen R, et al. Hepatology. 2009 Feb;49(2):692-4. doi: 10.1002/hep.22800. Hepatology. 2009. PMID: 19177565 No abstract available. - Genome-wide association studies reach hepatology.
Karlsen TH. Karlsen TH. J Hepatol. 2009 Jun;50(6):1278-80. doi: 10.1016/j.jhep.2009.03.002. Epub 2009 Mar 20. J Hepatol. 2009. PMID: 19395112 No abstract available.
Similar articles
- The PNPLA3 I148M polymorphism is associated with insulin resistance and nonalcoholic fatty liver disease in a normoglycaemic population.
Wang CW, Lin HY, Shin SJ, Yu ML, Lin ZY, Dai CY, Huang JF, Chen SC, Li SS, Chuang WL. Wang CW, et al. Liver Int. 2011 Oct;31(9):1326-31. doi: 10.1111/j.1478-3231.2011.02526.x. Epub 2011 Apr 5. Liver Int. 2011. PMID: 21745282 - Paradoxical dissociation between hepatic fat content and de novo lipogenesis due to PNPLA3 sequence variant.
Mancina RM, Matikainen N, Maglio C, Söderlund S, Lundbom N, Hakkarainen A, Rametta R, Mozzi E, Fargion S, Valenti L, Romeo S, Taskinen MR, Borén J. Mancina RM, et al. J Clin Endocrinol Metab. 2015 May;100(5):E821-5. doi: 10.1210/jc.2014-4464. Epub 2015 Mar 12. J Clin Endocrinol Metab. 2015. PMID: 25763607 - Characterization of European ancestry nonalcoholic fatty liver disease-associated variants in individuals of African and Hispanic descent.
Palmer ND, Musani SK, Yerges-Armstrong LM, Feitosa MF, Bielak LF, Hernaez R, Kahali B, Carr JJ, Harris TB, Jhun MA, Kardia SL, Langefeld CD, Mosley TH Jr, Norris JM, Smith AV, Taylor HA, Wagenknecht LE, Liu J, Borecki IB, Peyser PA, Speliotes EK. Palmer ND, et al. Hepatology. 2013 Sep;58(3):966-75. doi: 10.1002/hep.26440. Epub 2013 Jul 16. Hepatology. 2013. PMID: 23564467 Free PMC article. - PNPLA3-associated steatohepatitis: toward a gene-based classification of fatty liver disease.
Krawczyk M, Portincasa P, Lammert F. Krawczyk M, et al. Semin Liver Dis. 2013 Nov;33(4):369-79. doi: 10.1055/s-0033-1358525. Epub 2013 Nov 12. Semin Liver Dis. 2013. PMID: 24222094 Review. - PNPLA3 I148M variant in nonalcoholic fatty liver disease: demographic and ethnic characteristics and the role of the variant in nonalcoholic fatty liver fibrosis.
Chen LZ, Xin YN, Geng N, Jiang M, Zhang DD, Xuan SY. Chen LZ, et al. World J Gastroenterol. 2015 Jan 21;21(3):794-802. doi: 10.3748/wjg.v21.i3.794. World J Gastroenterol. 2015. PMID: 25624712 Free PMC article. Review.
Cited by
- Deciphering the multifaceted role of microRNAs in hepatocellular carcinoma: Integrating literature review and bioinformatics analysis for therapeutic insights.
Rahdan F, Saberi A, Saraygord-Afshari N, Hadizadeh M, Fayeghi T, Ghanbari E, Dianat-Moghadam H, Alizadeh E. Rahdan F, et al. Heliyon. 2024 Oct 18;10(20):e39489. doi: 10.1016/j.heliyon.2024.e39489. eCollection 2024 Oct 30. Heliyon. 2024. PMID: 39498055 Free PMC article. Review. - Genetic predisposition similarities between NASH and ASH: Identification of new therapeutic targets.
Bianco C, Casirati E, Malvestiti F, Valenti L. Bianco C, et al. JHEP Rep. 2021 Mar 30;3(3):100284. doi: 10.1016/j.jhepr.2021.100284. eCollection 2021 Jun. JHEP Rep. 2021. PMID: 34027340 Free PMC article. Review. - Coronavirus disease 2019 severity in obesity: Metabolic dysfunction-associated fatty liver disease in the spotlight.
Vasques-Monteiro IML, Souza-Mello V. Vasques-Monteiro IML, et al. World J Gastroenterol. 2021 Apr 28;27(16):1738-1750. doi: 10.3748/wjg.v27.i16.1738. World J Gastroenterol. 2021. PMID: 33967554 Free PMC article. Review. - A human liver chimeric mouse model for non-alcoholic fatty liver disease.
Bissig-Choisat B, Alves-Bezerra M, Zorman B, Ochsner SA, Barzi M, Legras X, Yang D, Borowiak M, Dean AM, York RB, Galvan NTN, Goss J, Lagor WR, Moore DD, Cohen DE, McKenna NJ, Sumazin P, Bissig KD. Bissig-Choisat B, et al. JHEP Rep. 2021 Mar 21;3(3):100281. doi: 10.1016/j.jhepr.2021.100281. eCollection 2021 Jun. JHEP Rep. 2021. PMID: 34036256 Free PMC article. - Dysfunctional VLDL metabolism in MASLD.
Marigorta UM, Millet O, Lu SC, Mato JM. Marigorta UM, et al. NPJ Metab Health Dis. 2024;2(1):16. doi: 10.1038/s44324-024-00018-1. Epub 2024 Jul 22. NPJ Metab Health Dis. 2024. PMID: 39049993 Free PMC article. Review.
References
- Browning JD, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–95. - PubMed
- McCullough AJ. The clinical features, diagnosis and natural history of nonalcoholic fatty liver disease. Clin Liver Dis. 2004;8:521–33. viii. - PubMed
- Adams LA, et al. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129:113–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RL1 HL092550-02/HL/NHLBI NIH HHS/United States
- P01 HL020948/HL/NHLBI NIH HHS/United States
- HL-20948/HL/NHLBI NIH HHS/United States
- HL-066681/HL/NHLBI NIH HHS/United States
- P01 HL020948-317281/HL/NHLBI NIH HHS/United States
- RL1 HL-092550/HL/NHLBI NIH HHS/United States
- RL1 HL092550/HL/NHLBI NIH HHS/United States
- 1PL1 DK081182/DK/NIDDK NIH HHS/United States
- U01 HL066681/HL/NHLBI NIH HHS/United States
- PL1 DK081182/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases