Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis - PubMed (original) (raw)
Review
Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis
Iuliana Ionita-Laza et al. Genomics. 2009 Jan.
Abstract
Structural genetic variation, including copy-number variation (CNV), constitutes a substantial fraction of total genetic variability and the importance of structural genetic variants in modulating human disease is increasingly being recognized. Early successes in identifying disease-associated CNVs via a candidate gene approach mandate that future disease association studies need to include structural genetic variation. Such analyses should not rely on previously developed methodologies that were designed to evaluate single nucleotide polymorphisms (SNPs). Instead, development of novel technical, statistical, and epidemiologic methods will be necessary to optimally capture this newly-appreciated form of genetic variation in a meaningful manner.
Figures
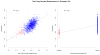
Figure 1. Raw copy number measurements vs. CNV calls for a CNV showing a continuous distribution
Shown in this figure, on the left side, is the association between a normally distributed, simulated phenotype and the intensity measurements at a single SNP position within a known copy-number variable region on chromosome 21 using a dataset of approximately 1200 individuals (CAMP study [53]). On the right side, the association between the same phenotype and the CNV calls (loss/no change) is shown. Losses were detected using a local false discovery rate (locFDR) approach [54], applied to the intensity measurements. As can be observed, for CNVs showing a continuous intensity distribution, forcibly classifying the raw measurements into discrete calls may result in loss of power compared to the original measurements, as illustrated by the drop in the R^2 value (the square of the correlation coefficient).
Similar articles
- What a difference copy number variation makes.
Kehrer-Sawatzki H. Kehrer-Sawatzki H. Bioessays. 2007 Apr;29(4):311-3. doi: 10.1002/bies.20554. Bioessays. 2007. PMID: 17373652 - On the analysis of copy-number variations in genome-wide association studies: a translation of the family-based association test.
Ionita-Laza I, Perry GH, Raby BA, Klanderman B, Lee C, Laird NM, Weiss ST, Lange C. Ionita-Laza I, et al. Genet Epidemiol. 2008 Apr;32(3):273-84. doi: 10.1002/gepi.20302. Genet Epidemiol. 2008. PMID: 18228561 - Genomic copy number variation, human health, and disease.
Wain LV, Armour JA, Tobin MD. Wain LV, et al. Lancet. 2009 Jul 25;374(9686):340-50. doi: 10.1016/S0140-6736(09)60249-X. Epub 2009 Jun 15. Lancet. 2009. PMID: 19535135 Review. - Clinical significance of germline copy number variation in susceptibility of human diseases.
Hu L, Yao X, Huang H, Guo Z, Cheng X, Xu Y, Shen Y, Xu B, Li D. Hu L, et al. J Genet Genomics. 2018 Jan 20;45(1):3-12. doi: 10.1016/j.jgg.2018.01.001. Epub 2018 Jan 5. J Genet Genomics. 2018. PMID: 29396143 Review. - Copy number polymorphisms and anticancer pharmacogenomics.
Gamazon ER, Huang RS, Dolan ME, Cox NJ. Gamazon ER, et al. Genome Biol. 2011;12(5):R46. doi: 10.1186/gb-2011-12-5-r46. Epub 2011 May 25. Genome Biol. 2011. PMID: 21609475 Free PMC article.
Cited by
- Genome-wide detection of copy number variations using high-density SNP genotyping platforms in Holsteins.
Jiang L, Jiang J, Yang J, Liu X, Wang J, Wang H, Ding X, Liu J, Zhang Q. Jiang L, et al. BMC Genomics. 2013 Feb 27;14:131. doi: 10.1186/1471-2164-14-131. BMC Genomics. 2013. PMID: 23442346 Free PMC article. - Genomic imbalance determines positive and negative modulation of gene expression in diploid maize.
Shi X, Yang H, Chen C, Hou J, Hanson KM, Albert PS, Ji T, Cheng J, Birchler JA. Shi X, et al. Plant Cell. 2021 May 31;33(4):917-939. doi: 10.1093/plcell/koab030. Plant Cell. 2021. PMID: 33677584 Free PMC article. - Trisomy 21 and facial developmental instability.
Starbuck JM, Cole TM 3rd, Reeves RH, Richtsmeier JT. Starbuck JM, et al. Am J Phys Anthropol. 2013 May;151(1):49-57. doi: 10.1002/ajpa.22255. Epub 2013 Mar 15. Am J Phys Anthropol. 2013. PMID: 23505010 Free PMC article. - Copy Number Variation and Osteoporosis.
Lovšin N. Lovšin N. Curr Osteoporos Rep. 2023 Apr;21(2):167-172. doi: 10.1007/s11914-023-00773-y. Epub 2023 Feb 16. Curr Osteoporos Rep. 2023. PMID: 36795294 Free PMC article. Review. - Accounting for uncertainty when assessing association between copy number and disease: a latent class model.
González JR, Subirana I, Escaramís G, Peraza S, Cáceres A, Estivill X, Armengol L. González JR, et al. BMC Bioinformatics. 2009 Jun 6;10:172. doi: 10.1186/1471-2105-10-172. BMC Bioinformatics. 2009. PMID: 19500389 Free PMC article.
References
- Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–6. - PubMed
- Edwards AO, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–4. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources