A novel biochemical method to identify target genes of individual microRNAs: identification of a new Caenorhabditis elegans let-7 target - PubMed (original) (raw)
A novel biochemical method to identify target genes of individual microRNAs: identification of a new Caenorhabditis elegans let-7 target
Yoshiki Andachi. RNA. 2008 Nov.
Abstract
MicroRNAs (miRNAs) are roughly 22-nucleotide regulatory RNAs that play important roles in many developmental and physiological processes. Animal miRNAs down-regulate target genes by forming imperfect base pairs with 3' untranslated regions (3' UTRs) of their mRNAs. Thousands of miRNAs have been discovered in several organisms. However, the target genes of almost all of these miRNAs remain to be identified. Here, we describe a method for isolating cDNA clones of target mRNAs that form base pairs in vivo with an endogenous miRNA of interest, in which the cDNAs are synthesized from the mRNAs using the miRNA as a reverse-transcription primer. The application of this method to Caenorhabditis elegans miRNA lin-4 under test conditions yielded many clones of the known target gene lin-14 that correspond to partial sequences 5' to lin-4 binding sites in the 3' UTR. The method was also applied to C. elegans miRNA let-7 and a new target gene responsible for the lethal phenotype in let-7 mutants was identified. These results demonstrate that the method is a useful way to identify targets on the basis of base pairing with individual miRNAs.
Figures
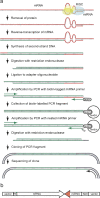
FIGURE 1.
Schematic of the method for isolating cDNA clones of mRNAs with which an endogenous miRNA forms base pairs. (a) Step-by-step description of the procedure. RNA and DNA strands are depicted in red and green, respectively. Worms were homogenized to prepare cytoplasmic extract. The extract was mixed with a detergent to destabilize proteins and incubated in a reverse-transcription reaction buffer to synthesize a first-strand cDNA using the miRNA as a primer. Polynucleotides collected from the buffer were incubated in a second-strand synthesis reaction buffer to synthesize DNA complementary to both the first-strand cDNA and the miRNA. After digestion of the double-stranded cDNA with a restriction endonuclease, the cDNA fragments were ligated to an adaptor oligonucleotide and then amplified by PCR with an adapter PCR primer and a biotin-tagged miRNA PCR primer corresponding to a partial sequence of the miRNA. Biotin-labeled PCR fragments were collected by binding to avidin beads and amplified by PCR with an adapter PCR primer and a nested miRNA PCR primer containing the NotI recognition sequence. Amplification fragments were digested with the same restriction endonuclease and NotI, and cloned into a vector. The sequences of inserts in randomly selected clones were determined with a vector primer. (b) Structure of a clone derived from a cDNA synthesized by reverse-transcription using a miRNA as a primer. The 5′-to-3′ directions of the mRNA and the miRNA sequences are indicated by arrows. The 3′-end of the miRNA excluded from the miRNA PCR primer is drawn in red.
FIGURE 2.
lin-14 3′ UTR clones isolated to identify lin-4 targets. Schematic of the lin-14 3′ UTR is depicted at the top where a HaeII recognition site is indicated with an arrow and lin-4 binding sites are shown by open triangles (▽). Partial sequence of the lin-14 3′ UTR is written where the lin-4 binding sites are surrounded by rectangles. The 3′ termini of cloned sequences are indicated by horizontal arrows. The numeral shows the number of cDNA libraries from which at least one clone corresponding to the arrow was isolated and the total number of clones is in parentheses. Note that no other lin-14 3′ UTR clones were isolated.
FIGURE 3.
Gene expression and orthologs of K10C3.4. (a) Coding regions and UTRs are drawn in green and red, respectively. The structures of plasmid constructs used in this study are also shown. (b_–_d) The expression of a GFP reporter containing the entire promoter (b), upstream promoter (c), and downstream promoter (d). (e,f) The localization of a GFP fusion protein in vulval cells (e) and in seam cells (f) of the same animal. All worms are at the L4 stage and the anterior is to the left. (g) Amino acid identities between orthologs. (Ce) C. elegans; (Cb) C. briggsae; (Cr) C. remanei; (Hs) Homo sapiens Transmembrane protein 98. Transmembrane domains are shown in black. (h) Alignment of 3′ UTR sequences. Numbers represent the _left_most nucleotides in base pairs downstream of the start of the 3′ UTRs. Conserved sequences complementary to the let-7 seed are surrounded by large rectangles. A nonconserved complementary sequence is denoted by a small hashed rectangle. Nucleotide substitutions are shown on the sequences along with the site number. An arrow indicates the 3′ terminus of the cloned sequence.
FIGURE 4.
Down-regulation of K10C3.4 and involvement of K10C3.4 in the let-7 vulval bursting phenotype. (a –d) Expression of a lacZ construct containing unc-54 3′ UTR (a,b) and K10C3.4 3′ UTR (c,d). Photos show GFP fluorescence (a,c) and bright field (b,d) images. Both worms are at the L4 stage and the anterior is to the left. Note that lacZ expression is repressed except in several head and tail cells in (d). (e) Frequency of lacZ expression among eight transgenic lines with 20 or more worms per line. (f– i) Bursting through the vulva in a K10C3.4 transgenic line containing the unc-54 3′ UTR at the young-adult stage (f) and at the gravid-adult stage (g), and in let-7(mn112) unc-3(e151) (h) and in let-7(mn112) unc-3(e151); K10C3.4(RNAi) (i). Note that many fertilized eggs were generated in i. (j) Frequency of vulval bursting in K10C3.4 transgenic worms among eight transgenic lines with 100 or more worms per line. (k) Frequency of survival at the young-adult stage in let-7(mn112) unc-3(e151) subjected to RNAi in five separate tests with 100 or more worms per test. Bars are mean ± SD, and a two-tailed, Student's _t_-test was used to determine significance (e,j,k).
Similar articles
- Caenorhabditis elegans period homolog lin-42 regulates the timing of heterochronic miRNA expression.
McCulloch KA, Rougvie AE. McCulloch KA, et al. Proc Natl Acad Sci U S A. 2014 Oct 28;111(43):15450-5. doi: 10.1073/pnas.1414856111. Epub 2014 Oct 15. Proc Natl Acad Sci U S A. 2014. PMID: 25319259 Free PMC article. - Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation.
Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Bagga S, et al. Cell. 2005 Aug 26;122(4):553-63. doi: 10.1016/j.cell.2005.07.031. Cell. 2005. PMID: 16122423 - Computational analysis of microRNA targets in Caenorhabditis elegans.
Watanabe Y, Yachie N, Numata K, Saito R, Kanai A, Tomita M. Watanabe Y, et al. Gene. 2006 Jan 3;365:2-10. doi: 10.1016/j.gene.2005.09.035. Epub 2005 Dec 13. Gene. 2006. PMID: 16356665 - C. elegans microRNAs.
Vella MC, Slack FJ. Vella MC, et al. WormBook. 2005 Sep 21:1-9. doi: 10.1895/wormbook.1.26.1. WormBook. 2005. PMID: 18050425 Free PMC article. Review. - Translational control of endogenous microRNA target genes in C. elegans.
Hurschler BA, Ding XC, Grosshans H. Hurschler BA, et al. Prog Mol Subcell Biol. 2010;50:21-40. doi: 10.1007/978-3-642-03103-8_2. Prog Mol Subcell Biol. 2010. PMID: 19841879 Review.
Cited by
- Novel miR390-dependent transacting siRNA precursors in plants revealed by a PCR-based experimental approach and database analysis.
Krasnikova MS, Milyutina IA, Bobrova VK, Ozerova LV, Troitsky AV, Solovyev AG, Morozov SY. Krasnikova MS, et al. J Biomed Biotechnol. 2009;2009:952304. doi: 10.1155/2009/952304. Epub 2009 Oct 13. J Biomed Biotechnol. 2009. PMID: 19859540 Free PMC article. - A whole-mount in situ hybridization method for microRNA detection in Caenorhabditis elegans.
Andachi Y, Kohara Y. Andachi Y, et al. RNA. 2016 Jul;22(7):1099-106. doi: 10.1261/rna.054239.115. Epub 2016 May 6. RNA. 2016. PMID: 27154969 Free PMC article. - let-7 coordinates the transition to adulthood through a single primary and four secondary targets.
Aeschimann F, Neagu A, Rausch M, Großhans H. Aeschimann F, et al. Life Sci Alliance. 2019 Mar 25;2(2):e201900335. doi: 10.26508/lsa.201900335. Print 2019 Apr. Life Sci Alliance. 2019. PMID: 30910805 Free PMC article. - Specific sequence determinants of miR-15/107 microRNA gene group targets.
Nelson PT, Wang WX, Mao G, Wilfred BR, Xie K, Jennings MH, Gao Z, Wang X. Nelson PT, et al. Nucleic Acids Res. 2011 Oct;39(18):8163-72. doi: 10.1093/nar/gkr532. Epub 2011 Jun 30. Nucleic Acids Res. 2011. PMID: 21724616 Free PMC article. - Survey of Computational Algorithms for MicroRNA Target Prediction.
Yue D, Liu H, Huang Y. Yue D, et al. Curr Genomics. 2009 Nov;10(7):478-92. doi: 10.2174/138920209789208219. Curr Genomics. 2009. PMID: 20436875 Free PMC article.
References
- Abrahante J.E., Daul A.L., Li M., Volk M.L., Tennessen J.M., Miller E.A., Rougvie A.E. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell. 2003;4:625–637. - PubMed
- Ambros V., Lee R.C., Lavanway A., Williams P.T., Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans . Curr. Biol. 2003;13:807–818. - PubMed
- Andachi Y. Caenorhabditis elegans T-box genes tbx-9 and tbx-8 are required for formation of hypodermis and body-wall muscle in embryogenesis. Genes Cells. 2004;9:331–344. - PubMed
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. - PubMed
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources