Delta-like 4 is indispensable in thymic environment specific for T cell development - PubMed (original) (raw)
Comparative Study
. 2008 Oct 27;205(11):2507-13.
doi: 10.1084/jem.20080134. Epub 2008 Sep 29.
Affiliations
- PMID: 18824583
- PMCID: PMC2571926
- DOI: 10.1084/jem.20080134
Comparative Study
Delta-like 4 is indispensable in thymic environment specific for T cell development
Katsuto Hozumi et al. J Exp Med. 2008.
Abstract
The thymic microenvironment is required for T cell development in vivo. However, in vitro studies have shown that when hematopoietic progenitors acquire Notch signaling via Delta-like (Dll)1 or Dll4, they differentiate into the T cell lineage in the absence of a thymic microenvironment. It is not clear, however, whether the thymus supports T cell development specifically by providing Notch signaling. To address this issue, we generated mice with a loxP-flanked allele of Dll4 and induced gene deletion specifically in thymic epithelial cells (TECs). In the thymus of mutant mice, the expression of Dll4 was abrogated on the epithelium, and the proportion of hematopoietic cells bearing the intracellular fragment of Notch1 (ICN1) was markedly decreased. Corresponding to this, CD4 CD8 double-positive or single-positive T cells were not detected in the thymus. Further analysis showed that the double-negative cell fraction was lacking T cell progenitors. The enforced expression of ICN1 in hematopoietic progenitors restored thymic T cell differentiation, even when the TECs were deficient in Dll4. These results indicate that the thymus-specific environment for determining T cell fate indispensably requires Dll4 expression to induce Notch signaling in the thymic immigrant cells.
Figures
Figure 1.
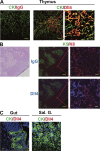
Dll4 was expressed on thymic epithelium. The expression of Dll4 on the epithelium was detected in thymus (A and B, Thymus), but not gut (C, Gut) or salivary gland (C, Sal. G.) by immunofluorescence histochemistry. Staining was done with anti–pan-cytokeratin (CK; A, green), anti-K5 (K5; B, green), anti-K8 (K8; B, red), anti-Dll4 (Dll4; A, red; B, blue), or control IgG (IgG; A, red; B, blue) antibodies and analyzed by confocal laser microscopy. (A and B, right) High-magnification images of the red square in the middle images. (B, left) Hemotoxylin and eosin staining of serial section. Bars: (A, left and middle) 50 μm; (A, right) 10 μm; (B, left) 200 μm; (B, right) 10 μm; (C) 20 μm.
Figure 2.
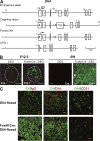
Specific abrogation of Dll4 on thymic epithelium. (A) Targeted insertion of loxP sequences flanking a part of exon 1 and whole exons 2 and 3 of the Dll4 gene. Numbers indicate exon number. 1*, first exon modified with loxP sequence; 1**, part of exon 1 after gene deletion; ATG, translational initiation codon of Dll4; triangles, loxP sequences; open boxes, Dll4 exons; B, BglII; C, ClaI; RI, EcoRI; Xb, XbaI; Xh, XhoI. (B) Cre activity was targeted to thymic epithelia using mice, designated FoxN1-Cre, in which this recombinase is under the transcriptional control of the FoxN1 locus. The timing and specificity of Cre-mediated recombination was visualized by the expression of enhanced GFP (eGFP) in thymus tissue sections of E12.5 (left) and 6-wk-old (right) crosses of FoxN1-Cre mice with the transgenic Z/EG reporter mice. The thymus anlage in the left image is outlined with a dashed line. C, cortex; M, medulla. (C) The expression of Dll4 on thymic epithelial, but not endothelial, cells was abrogated in _Dll4_lox/loxFoxN1-Cre (FoxN1Cre:Dll4-floxed) mice. Epithelial and endothelial cells in the thymus were identified by the expression of cytokeratin (CK, green, left and middle) and CD31 (red, right), respectively, with Dll4 (red, middle; green, right). Bars: (B and C) 50 μm.
Figure 3.
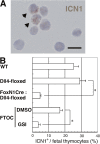
Notch signaling was decreased in the thymus with Dll4-null epithelial cells. (A) The cleaved Notch1 fragment was found in fetal thymocytes of E15.5 _Dll4_lox/lox (Dll4-floxed) embryos. The cells with cleaved Notch1 are indicated with arrowheads. Bar, 10 μm. (B) Frequencies of cells with cleaved Notch1 (ICN1+; mean percentage from five fields in a slide with >100 cells from each embryo ± SD) were counted in E15.5 WT (n = 3), _Dll4_lox/loxFoxN1Cre (FoxN1Cre:Dll4-floxed, n = 6), or _Dll4_lox/lox (Dll4-floxed, n = 6) mice. The cultured DN cells were prepared after the FTOC of E14.5 WT fetal thymus for 4 d with γ-secretase inhibitor (GSI, n = 3) or without γ-secretase inhibitor (DMSO, n = 3). Asterisk indicates unpaired Student's t test; P < 0.001.
Figure 4.
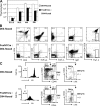
No T cells, but substantial B cells, appear in the thymus with Dll4-null epithelium. (A) Thymic cellularity (mean ± SD) of fetus (E15.5; left, n = 3; right, n = 5), neonate (N.B.; left, n = 5; right, n = 3), 4-wk-old (4W, n = 4), or 8-wk-old (8W, n = 3) _Dll4_lox/loxFoxN1-Cre (FoxN1Cre:Dll4-floxed, shaded columns) mice compared with _Dll4_lox/lox (Dll4-floxed, open columns) mice. Asterisks indicate unpaired Student's t test; P < 0.001. (B) Flow cytometry of thymocytes from young adult (4W) _Dll4_lox/lox mice with (bottom, FoxN1Cre:Dll4-floxed) or without (top, Dll4-floxed) FoxN1-Cre transgene. Thymocytes were stained with monoclonal antibodies to surface molecules as shown. Numbers in the profiles indicate the relative percentages, in CD4−CD8− cells (right, CD4+CD8 vs. Thy1.2), for each corresponding quadrant or fraction. (C) The ETP population was not observed in _Dll4_lox/loxFoxN1-Cre (4W) mice. Thymocytes from _Dll4_lox/lox (top) or _Dll4_lox/loxFoxN1-Cre mice (bottom) were stained for lineage markers (Lin.: CD4, CD8, CD3, B220, CD19, Gr1, CD11b, TER119), CD44, CD25, CD24, and c-kit. Profiles are shown with the gate (Lin.−, middle; Lin.−CD44+CD25− as DN1, right). Numbers on the plots represent the frequency of cells lying in the indicated regions within the gate. The ETP population is identified as Lin.−CD44+CD25−CD24− or lowc-kit+ cells (DN1a+b). DN1c and DN1–3 populations are also shown in the plots. Data are representative of three experiments.
Figure 5.
Hematopoietic progenitors with active form of Notch1 give rise to T cell lineage in Dll4-deficient thymus. Hematopoietic progenitors (Lin.−c-kit+ cells) derived from E15.5 fetal liver were infected with the retrovirus encoding the intracellular region of Notch1 (ICN1) or empty vector (Mock). Infected cells were monitored by expression of GFP. These cells were cultured into deoxyguanosine-treated thymic lobes of _Dll4_lox/loxFoxN1-Cre (FoxN1Cre:Dll4-floxed) or _Dll4_lox/lox (Dll4-floxed) fetuses for 13 d as FTOC and analyzed for the expression of CD4, CD8, CD25, and CD19 (A). Alternatively, these cells were injected intravenously into irradiated _Dll4_lox/loxFoxN1-Cre (FoxN1Cre:Dll4-floxed) or _Dll4_lox/lox (Dll4-floxed) neonate mice. After 3 wk, thymocytes were analyzed for expression of CD4, CD8, and CD19 (B). Profiles are shown with the gate (GFP+ or GFP+CD4−CD8−, DN-gated in A). Numbers in the profiles indicate the relative percentages for each corresponding quadrant.
Similar articles
- Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment.
Koch U, Fiorini E, Benedito R, Besseyrias V, Schuster-Gossler K, Pierres M, Manley NR, Duarte A, Macdonald HR, Radtke F. Koch U, et al. J Exp Med. 2008 Oct 27;205(11):2515-23. doi: 10.1084/jem.20080829. Epub 2008 Sep 29. J Exp Med. 2008. PMID: 18824585 Free PMC article. - Ongoing Dll4-Notch signaling is required for T-cell homeostasis in the adult thymus.
Billiard F, Kirshner JR, Tait M, Danave A, Taheri S, Zhang W, Waite JC, Olson K, Chen G, Coetzee S, Hylton D, Murphy AJ, Yancopoulos GD, Thurston G, Skokos D. Billiard F, et al. Eur J Immunol. 2011 Aug;41(8):2207-16. doi: 10.1002/eji.201041343. Epub 2011 Jul 4. Eur J Immunol. 2011. PMID: 21598246 - Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions.
Schmitt TM, Ciofani M, Petrie HT, Zúñiga-Pflücker JC. Schmitt TM, et al. J Exp Med. 2004 Aug 16;200(4):469-79. doi: 10.1084/jem.20040394. J Exp Med. 2004. PMID: 15314075 Free PMC article. - Regulation of Notch signaling during T- and B-cell development by O-fucose glycans.
Stanley P, Guidos CJ. Stanley P, et al. Immunol Rev. 2009 Jul;230(1):201-15. doi: 10.1111/j.1600-065X.2009.00791.x. Immunol Rev. 2009. PMID: 19594638 Review. - Gammadelta T cell development--having the strength to get there.
Pennington DJ, Silva-Santos B, Hayday AC. Pennington DJ, et al. Curr Opin Immunol. 2005 Apr;17(2):108-15. doi: 10.1016/j.coi.2005.01.009. Curr Opin Immunol. 2005. PMID: 15766668 Review.
Cited by
- The Earliest T-Precursors in the Mouse Embryo Are Susceptible to Leukemic Transformation.
Ding J, Cardoso AA, Yoshimoto M, Kobayashi M. Ding J, et al. Front Cell Dev Biol. 2021 Apr 29;9:634151. doi: 10.3389/fcell.2021.634151. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33996794 Free PMC article. - Intrathymic Selection and Defects in the Thymic Epithelial Cell Development.
García-Ceca J, Montero-Herradón S, Zapata AG. García-Ceca J, et al. Cells. 2020 Oct 2;9(10):2226. doi: 10.3390/cells9102226. Cells. 2020. PMID: 33023072 Free PMC article. Review. - Loss of T-cell quiescence by targeting Slfn2 prevents the development and progression of T-ALL.
Goldshtein A, Zerbib SM, Omar I, Cohen-Daniel L, Popkin D, Berger M. Goldshtein A, et al. Oncotarget. 2016 Jul 26;7(30):46835-46847. doi: 10.18632/oncotarget.9390. Oncotarget. 2016. PMID: 27206675 Free PMC article. - Notch2 complements Notch1 to mediate inductive signaling that initiates early T cell development.
Romero-Wolf M, Shin B, Zhou W, Koizumi M, Rothenberg EV, Hosokawa H. Romero-Wolf M, et al. J Cell Biol. 2020 Oct 5;219(10):e202005093. doi: 10.1083/jcb.202005093. J Cell Biol. 2020. PMID: 32756905 Free PMC article. - Notch signaling in hematopoietic cell transplantation and T cell alloimmunity.
Ebens CL, Maillard I. Ebens CL, et al. Blood Rev. 2013 Nov;27(6):269-77. doi: 10.1016/j.blre.2013.08.001. Epub 2013 Aug 31. Blood Rev. 2013. PMID: 24050990 Free PMC article. Review.
References
- Anderson, G., and E.J. Jenkinson. 2001. Lymphostromal interactions in thymic development and function. Nat. Rev. Immunol. 1:31–40. - PubMed
- Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H.R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558. - PubMed
- Han, H., K. Tanigaki, N. Yamamoto, K. Kuroda, M. Yoshimoto, T. Nakahata, K. Ikuta, and T. Honjo. 2002. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14:637–645. - PubMed
- Pui, J.C., D. Allman, L. Xu, S. DeRocco, F.G. Karnell, S. Bakkour, J.Y. Lee, T. Kadesch, R.R. Hardy, J.C. Aster, and W.S. Pear. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 11:299–308. - PubMed
- Hozumi, K., N. Abe, S. Chiba, H. Hirai, and S. Habu. 2003. Active form of Notch members can enforce T lymphopoiesis on lymphoid progenitors in the monolayer culture specific for B cell development. J. Immunol. 170:4973–4979. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous