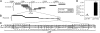Caulobacter requires a dedicated mechanism to initiate chromosome segregation - PubMed (original) (raw)
Caulobacter requires a dedicated mechanism to initiate chromosome segregation
Esteban Toro et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Chromosome segregation in bacteria is rapid and directed, but the mechanisms responsible for this movement are still unclear. We show that Caulobacter crescentus makes use of and requires a dedicated mechanism to initiate chromosome segregation. Caulobacter has a single circular chromosome whose origin of replication is positioned at one cell pole. Upon initiation of replication, an 8-kb region of the chromosome containing both the origin and parS moves rapidly to the opposite pole. This movement requires the highly conserved ParABS locus that is essential in Caulobacter. We use chromosomal inversions and in vivo time-lapse imaging to show that parS is the Caulobacter site of force exertion, independent of its position in the chromosome. When parS is moved farther from the origin, the cell waits for parS to be replicated before segregation can begin. Also, a mutation in the ATPase domain of ParA halts segregation without affecting replication initiation. Chromosome segregation in Caulobacter cannot occur unless a dedicated parS guiding mechanism initiates movement.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Extra copies of parS DNA impair cell viability. (A and B) DNA-copy-number screen. Cosmids (A) or plasmids (B), carrying an antibiotic resistance cassette and a region (lines) of the Caulobacter genome, were transformed into cells. Solid lines represent constructs that permitted growth of colonies on selective plates; dashed lines represent constructs that did not allow growth. (C) Sequence of the wild-type parS site (WT) and parS with mutated ParB-binding motifs (Mut). Asterisks denote the location of base-pair changes in parS. (D) Transformation efficiency (relative number of colonies at day 3) of plasmids carrying either the wild-type (_parS_-WT) or mutated (_parS_-Mut) versions of parS. * = 0.003. Bars represent the means of seven separate experiments. Here and elsewhere, error bars represent standard error of the mean (SEM).
Fig. 2.
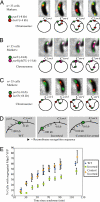
A 10-kb region including parS contains the site of force exertion during segregation. (A–C) Segregation pattern of different loci and accompanying schematics of the position of markers used and order of segregation. Note that although micrographs show only one cell, the segregation order shown (parS segregating before the other tagged locus) was repeated in all cells observed. In all cases, parS visualized with MipZ-YFP. For clarity, schematics are not to scale. (A) lacO inserted at +4 kb and visualized with LacI-CFP. (B) parS(pMT1) inserted at −5 kb and visualized with CFP-pMT1Δ23ParB. (C) lacO inserted at −64 kb and visualized with LacI-CFP. (D) Schematic of the chromosomal configuration of inversion strains constructed by site-specific recombination. (E) Separating parS from Cori delays segregation. Plotted are the percentage of cells with two distinct MipZ-YFP foci as a function of time from synchrony. To avoid phototoxicity effects, a new field of cells was imaged for each time point. Chromosome configurations as in D. Symbols represent means of three experiments.
Fig. 3.
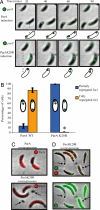
A mutation in ParA abrogates chromosome segregation and produces anucleate minicells. (A) Time-lapse fluorescence micrographs of CFP-ParB in synchronized cells undergoing segregation after 60 min of ParA-mCherry (Upper) or ParAK20R-mCherry (Lower) induction with 0.03% xylose. (B) Percentage of cells with two foci that were either partially (blue bars) or fully (yellow bars) segregated 90 min after synchrony, in cultures treated as in A. * = 0.5%. Bars represent the average of two independent experiments, each with at least 150 cells counted for each strain. (C) ParA-mCherry (Upper) and ParAK20R-mCherry (Lower) localization in cells induced for 5 hours with 0.3% xylose. Arrows point to minicells produced only with the mutant ParA. (D) Localization of ParAK20R-mCherry and CFP-ParB (Upper) and DAPI (Lower) in cells carrying ParAK20R-mCherry driven by the xylose promoter and treated as in C. Arrows point to minicells that show no CFP-ParB foci and very low DAPI fluorescence. Levels of DAPI fluorescence have been increased to aid visualization of the very low fluorescence in minicells.
Fig. 4.
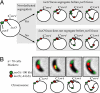
Replication is not sufficient to initiate chromosome segregation. (A) Schematic of the strain constructed to test for the presence of nondedicated segregation mechanisms, and possible outcomes. (Upper) Nondedicated segregation occurs. Replicated DNA is moved independently of the parABS mechanism, and movement of the lacO arrays takes place before parS duplication. (Lower) Nondedicated mechanisms are not able to initiate chromosome segregation, and lacO movement does not take place until after parS has begun segregating. (B) Results of the experiment outlined in panel A. parS was followed by using MipZ-YFP, lacO arrays were followed by using LacI-CFP. Seventy cells were observed segregating and in all cases parS segregated before the lacO arrays.
Fig. 5.
parS is fixed at the cell pole, whereas Cori is not. (A and B) Fluorescent micrographs, schematic, and quantification of the relative positions of parS and lacO in cells before and after inversion of the chromosome region shown. (A) lacO originally positioned at −64 kb. (B) lacO positioned at +4 kb. Bars represent the average of three independent experiments, each with at least 100 cells counted.
Similar articles
- Chromosome Dynamics in Bacteria: Triggering Replication at the Opposite Location and Segregation in the Opposite Direction.
Meléndez AB, Menikpurage IP, Mera PE. Meléndez AB, et al. mBio. 2019 Jul 30;10(4):e01002-19. doi: 10.1128/mBio.01002-19. mBio. 2019. PMID: 31363028 Free PMC article. - Compaction and transport properties of newly replicated Caulobacter crescentus DNA.
Hong SH, McAdams HH. Hong SH, et al. Mol Microbiol. 2011 Dec;82(6):1349-58. doi: 10.1111/j.1365-2958.2011.07899.x. Epub 2011 Nov 16. Mol Microbiol. 2011. PMID: 22085253 - Permissive zones for the centromere-binding protein ParB on the Caulobacter crescentus chromosome.
Tran NT, Stevenson CE, Som NF, Thanapipatsiri A, Jalal ASB, Le TBK. Tran NT, et al. Nucleic Acids Res. 2018 Feb 16;46(3):1196-1209. doi: 10.1093/nar/gkx1192. Nucleic Acids Res. 2018. PMID: 29186514 Free PMC article. - Control of chromosome replication in caulobacter crescentus.
Marczynski GT, Shapiro L. Marczynski GT, et al. Annu Rev Microbiol. 2002;56:625-56. doi: 10.1146/annurev.micro.56.012302.161103. Epub 2002 Jan 30. Annu Rev Microbiol. 2002. PMID: 12142494 Review. - Cell cycle regulation in Caulobacter: location, location, location.
Goley ED, Iniesta AA, Shapiro L. Goley ED, et al. J Cell Sci. 2007 Oct 15;120(Pt 20):3501-7. doi: 10.1242/jcs.005967. J Cell Sci. 2007. PMID: 17928306 Review.
Cited by
- Phospho-signaling couples polar asymmetry and proteolysis within a membraneless microdomain in Caulobacter crescentus.
Ahmed YM, Brown LM, Varga K, Bowman GR. Ahmed YM, et al. Nat Commun. 2024 Oct 28;15(1):9282. doi: 10.1038/s41467-024-53395-y. Nat Commun. 2024. PMID: 39468040 Free PMC article. - Molecular model of a bacterial flagellar motor in situ reveals a "parts-list" of protein adaptations to increase torque.
Drobnič T, Cohen EJ, Calcraft T, Alzheimer M, Froschauer K, Svensson S, Hoffmann WH, Singh N, Garg SG, Henderson L, Umrekar TR, Nans A, Ribardo D, Pedaci F, Nord AL, Hochberg GKA, Hendrixson DR, Sharma CM, Rosenthal PB, Beeby M. Drobnič T, et al. bioRxiv [Preprint]. 2024 Oct 9:2023.09.08.556779. doi: 10.1101/2023.09.08.556779. bioRxiv. 2024. PMID: 39416179 Free PMC article. Preprint. - Loop-extruders alter bacterial chromosome topology to direct entropic forces for segregation.
Harju J, van Teeseling MCF, Broedersz CP. Harju J, et al. Nat Commun. 2024 May 30;15(1):4618. doi: 10.1038/s41467-024-49039-w. Nat Commun. 2024. PMID: 38816445 Free PMC article. - Chromosome organization shapes replisome dynamics in Caulobacter crescentus.
Zhang C, Joseph AM, Casini L, Collier J, Badrinarayanan A, Manley S. Zhang C, et al. Nat Commun. 2024 Apr 24;15(1):3460. doi: 10.1038/s41467-024-47849-6. Nat Commun. 2024. PMID: 38658616 Free PMC article. - Three factors ParA, TipN, and DnaA-mediated chromosome replication initiation are contributors of centromere segregation in Caulobacter crescentus.
Letzkus M, Trela C, Mera PE. Letzkus M, et al. Mol Biol Cell. 2024 May 1;35(5):ar68. doi: 10.1091/mbc.E23-12-0503. Epub 2024 Apr 3. Mol Biol Cell. 2024. PMID: 38568781 Free PMC article.
References
- Jacob F, Brenner S, Cuzin F. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp Quant Biol. 1963;28:329–348.
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM051426/GM/NIGMS NIH HHS/United States
- R24 GM073011/GM/NIGMS NIH HHS/United States
- GM073011-04/GM/NIGMS NIH HHS/United States
- R01 GM51426 R24/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous