Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals - PubMed (original) (raw)
. 2008 Oct 30;455(7217):1193-7.
doi: 10.1038/nature07415. Epub 2008 Oct 1.
Affiliations
- PMID: 18830242
- PMCID: PMC3837422
- DOI: 10.1038/nature07415
Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals
Andrew Grimson et al. Nature. 2008.
Abstract
In bilaterian animals, such as humans, flies and worms, hundreds of microRNAs (miRNAs), some conserved throughout bilaterian evolution, collectively regulate a substantial fraction of the transcriptome. In addition to miRNAs, other bilaterian small RNAs, known as Piwi-interacting RNAs (piRNAs), protect the genome from transposons. Here we identify small RNAs from animal phyla that diverged before the emergence of the Bilateria. The cnidarian Nematostella vectensis (starlet sea anemone), a close relative to the Bilateria, possesses an extensive repertoire of miRNA genes, two classes of piRNAs and a complement of proteins specific to small-RNA biology comparable to that of humans. The poriferan Amphimedon queenslandica (sponge), one of the simplest animals and a distant relative of the Bilateria, also possesses miRNAs, both classes of piRNAs and a full complement of the small-RNA machinery. Animal miRNA evolution seems to have been relatively dynamic, with precursor sizes and mature miRNA sequences differing greatly between poriferans, cnidarians and bilaterians. Nonetheless, miRNAs and piRNAs have been available as classes of riboregulators to shape gene expression throughout the evolution and radiation of animal phyla.
Figures
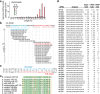
Figure 1. Phylogenetic distribution of annotated miRNAs
Cladogram of selected eukaryotes, with organisms investigated in this study indicated in red. Branching order of Bilateria is according to ref. 28 and the references therein, and that of basal Metazoa is according to ref. 17 (Supplementary Discussion). Annotated miRNA tallies are from miRBase (v10.1).
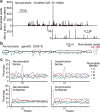
Figure 2. The miRNAs of N. vectensis
a, Length distribution of genome-matching sequencing reads representing small RNAs, plotted by 5′-nucleotide (nt) identity. Matches to ribosomal DNA were omitted. b, Sequencing reads matching the mir-2024d hairpin. The sequence of the mir-2024d hairpin is depicted above the bracket-notation of its predicted secondary structure. The sequenced small RNAs mapping to the hairpin are aligned below, with the number of reads shown on the left, and the designated miRNA and miRNA* species coloured red and blue, respectively. Analogous information is provided for the other newly identified miRNAs (Supplementary Data 1). c, Predicted secondary structure of the mir-2024d hairpin, indicating the miRNA and miRNA* species. d, The 40 Nematostella miRNAs. MicroRNA read counts include those sharing the dominant 5′ terminus but possessing variable 3′ termini. Occasionally the only sequenced miRNA* species corresponded to a variant miRNA species rather than the major species (counts in brackets). e, Alignment of miR-100 homologues (Danio rerio, D. rerio; Xenopus tropicalis, X. tropicalis).
Figure 3. The miRNAs of Amphimedon queenslandica
a, Length distribution of genome-matching sequencing reads representing small RNAs, plotted by 5′-nucleotide identity. Matches to ribosomal DNA were omitted. b, The Amphimedon miRNAs, shown as in Fig. 2d. Information analogous to that of Fig. 2b is provided for these miRNAs (Supplementary Data 2). c, Predicted secondary structure of the mir-2018 hairpin. d, Relative expression of Amphimedon miRNAs, as indicated by sequencing frequency from adult and embryo samples. e, Cumulative distributions of pre-miRNA lengths from miRNA transcripts of the species indicated. Amphimedon pre-miRNAs were significantly larger than those from any other animal species examined (P < 10−5, Wilcoxon rank-sum test), whereas those from Nematostella were significantly smaller (P < 10−5).
Figure 4. The piRNAs of basal metazoans
a, Distribution of reads matching a Nematostella piRNA locus. Plotted is the number of matching reads with 5′ nucleotide falling within each 100-nucleotide window (main graph) or at each nucleotide (higher-resolution inset) spanning the genomic region. Bars above and below the _x_-axis indicate matches to the indicated strand, with black bars indicating reads with a 5′-U and red bars indicating the sum of all other reads. For reads also matching other genomic loci, counts were normalized by total genome matches. Other annotated piRNA loci are presented in Supplementary Tables 3 and 4. kb, kilobases. b, An annotated pre-mRNA corresponding to numerous small RNAs resistant to periodate treatment. Annotated coding segments (open boxes) and intron segments (black line) are indicated. The gene was homologous to endonuclease/reverse transcriptases of other genomes and presumed to be a transposase. Small RNAs with unique 5′ ends are represented by coloured bars above or below the transcript (sense and antisense, respectively), with colours indicating the read numbers (normalized to account for the number of transcriptome matches). Small RNAs matching splice junctions (observed only for sense reads) are represented by discontinuous bars, linked by dashed lines. Other Nematostella and Amphimedon coding regions matching candidate piRNAs are listed in Supplementary Tables 3 and 4. c, Nucleotide composition of periodate-resistant small RNAs matching the indicated strand of Nematostella or Amphimedon annotated coding regions.
Comment in
- Evolutionary biology: Small regulatory RNAs pitch in.
Technau U. Technau U. Nature. 2008 Oct 30;455(7217):1184-5. doi: 10.1038/4551184a. Nature. 2008. PMID: 18972008 No abstract available.
Similar articles
- Diverse RNA interference strategies in early-branching metazoans.
Calcino AD, Fernandez-Valverde SL, Taft RJ, Degnan BM. Calcino AD, et al. BMC Evol Biol. 2018 Nov 1;18(1):160. doi: 10.1186/s12862-018-1274-2. BMC Evol Biol. 2018. PMID: 30382896 Free PMC article. - The methyltransferase HEN1 is required in Nematostella vectensis for microRNA and piRNA stability as well as larval metamorphosis.
Modepalli V, Fridrich A, Agron M, Moran Y. Modepalli V, et al. PLoS Genet. 2018 Aug 17;14(8):e1007590. doi: 10.1371/journal.pgen.1007590. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30118479 Free PMC article. - Elucidating the Small Regulatory RNA Repertoire of the Sea Anemone Anemonia viridis Based on Whole Genome and Small RNA Sequencing.
Urbarova I, Patel H, Forêt S, Karlsen BO, Jørgensen TE, Hall-Spencer JM, Johansen SD. Urbarova I, et al. Genome Biol Evol. 2018 Feb 1;10(2):410-426. doi: 10.1093/gbe/evy003. Genome Biol Evol. 2018. PMID: 29385567 Free PMC article. - Diverse small non-coding RNAs in RNA interference pathways.
Li L, Liu Y. Li L, et al. Methods Mol Biol. 2011;764:169-82. doi: 10.1007/978-1-61779-188-8_11. Methods Mol Biol. 2011. PMID: 21748640 Review. - Rising starlet: the starlet sea anemone, Nematostella vectensis.
Darling JA, Reitzel AR, Burton PM, Mazza ME, Ryan JF, Sullivan JC, Finnerty JR. Darling JA, et al. Bioessays. 2005 Feb;27(2):211-21. doi: 10.1002/bies.20181. Bioessays. 2005. PMID: 15666346 Review.
Cited by
- Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants.
Mukherjee K, Campos H, Kolaczkowski B. Mukherjee K, et al. Mol Biol Evol. 2013 Mar;30(3):627-41. doi: 10.1093/molbev/mss263. Epub 2012 Nov 22. Mol Biol Evol. 2013. PMID: 23180579 Free PMC article. - The origin of the ADAR gene family and animal RNA editing.
Grice LF, Degnan BM. Grice LF, et al. BMC Evol Biol. 2015 Jan 29;15(1):4. doi: 10.1186/s12862-015-0279-3. BMC Evol Biol. 2015. PMID: 25630791 Free PMC article. - Small RNAs from a Big Genome: The piRNA Pathway and Transposable Elements in the Salamander Species Desmognathus fuscus.
Madison-Villar MJ, Sun C, Lau NC, Settles ML, Mueller RL. Madison-Villar MJ, et al. J Mol Evol. 2016 Oct;83(3-4):126-136. doi: 10.1007/s00239-016-9759-3. Epub 2016 Oct 14. J Mol Evol. 2016. PMID: 27743003 - shutdown is a component of the Drosophila piRNA biogenesis machinery.
Preall JB, Czech B, Guzzardo PM, Muerdter F, Hannon GJ. Preall JB, et al. RNA. 2012 Aug;18(8):1446-57. doi: 10.1261/rna.034405.112. Epub 2012 Jul 2. RNA. 2012. PMID: 22753781 Free PMC article. - Transcriptome profiling of the demosponge Amphimedon queenslandica reveals genome-wide events that accompany major life cycle transitions.
Conaco C, Neveu P, Zhou H, Arcila ML, Degnan SM, Degnan BM, Kosik KS. Conaco C, et al. BMC Genomics. 2012 May 30;13:209. doi: 10.1186/1471-2164-13-209. BMC Genomics. 2012. PMID: 22646746 Free PMC article.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. - PubMed
- Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases