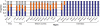Amerindian Helicobacter pylori strains go extinct, as european strains expand their host range - PubMed (original) (raw)
Amerindian Helicobacter pylori strains go extinct, as european strains expand their host range
Maria G Domínguez-Bello et al. PLoS One. 2008.
Abstract
We studied the diversity of bacteria and host in the H. pylori-human model. The human indigenous bacterium H. pylori diverged along with humans, into African, European, Asian and Amerindian groups. Of these, Amerindians have the least genetic diversity. Since niche diversity widens the sets of resources for colonizing species, we predicted that the Amerindian H. pylori strains would be the least diverse. We analyzed the multilocus sequence (7 housekeeping genes) of 131 strains: 19 cultured from Africans, 36 from Spanish, 11 from Koreans, 43 from Amerindians and 22 from South American Mestizos. We found that all strains that had been cultured from Africans were African strains (hpAfrica1), all from Spanish were European (hpEurope) and all from Koreans were hspEAsia but that Amerindians and Mestizos carried mixed strains: hspAmerind and hpEurope strains had been cultured from Amerindians and hpEurope and hpAfrica1 were cultured from Mestizos. The least genetically diverse H. pylori strains were hspAmerind. Strains hpEurope were the most diverse and showed remarkable multilocus sequence mosaicism (indicating recombination). The lower genetic structure in hpEurope strains is consistent with colonization of a diversity of hosts. If diversity is important for the success of H. pylori, then the low diversity of Amerindian strains might be linked to their apparent tendency to disappear. This suggests that Amerindian strains may lack the needed diversity to survive the diversity brought by non-Amerindian hosts.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Distribution of ABO blood groups in humans from Africa, Asia, Western countries and in Amerindians.
The blue, orange and grey bars represent respectively O, A and B allele frequencies –. The remarkable dominance of O blood groups among Amerindians from South America affects the ABO blood group recognition by H. pylori strains.
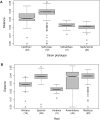
Figure 2. Pairwise genetic distances between H. pylori strains grouped by bacterial population (2A), or according to their human host (2B).
Differences in pairwise distances among strains in Fig. 2A were significant (Kruskal-Wallis test, p<2.2×10−16, Wilcoxon and Bonferroni; p<10−14) with a decreasing order _hpEurope_>_hpAfrica1_>_hpEastAsia_>hspAmerind. When grouped by the human host from which each strain was isolated (Fig 2B), strain diversity in Amerindians was as high as in Spanish and Mestizos (with no significant differences among them; Wilcoxon Pairwise comparison _p_>0.7), with a decreasing order Spaniards = Amerindians = Mestizos>Africans>Koreans. The waist of the dress-like box is the median with the waist side openings indicating the 95% interval for the median; the top represents the 3rd quartile and the bottom the 1st quartile. The interval in dashed lines represents a maximum of 1.5× interquartile range and the open circles are outliers. Permutation tests confirmed group differences in strain diversity.
Figure 3. Mosaic structure of the multilocus H. pylori sequences in representative strains.
The ancestral source of each polymorphic nucleotide is shown by a vertical line for each of the seven gene fragments in the multilocus analysis of 10 representative strains from each group (see the legend below Mestizo strains). Individual nucleotides were derived from ancestral Europe1 (grey), ancestral Europe2 (green), ancestral Africa1 (blue) and ancestral EastAsia (yellow). Nucleotides not assigned with >50% probability to any one population are indicated by white lines. African and European components can be observed in hpEurope strains from Spaniards and Mestizos, as well as in hpAfrica1 from Mestizos, while African hpAfrica1 strains and Amerindian hspAmerind strains tested were largely homogeneous.
Figure 4. Development of inter-strain genetic diversity over time as two populations recombine.
In the context of our hypothesis, a low diversity H. pylori population (P1) arose from co-evolution with the isolated Amerindian host population. With the introduction of new H. pylori strains (P2) the new population formed (P3) is more diverse than any of the source populations alone. Selection acts and strains recombine with the consequent homogenization of the population (P3′). The cycle is repeated when new populations (P4) arrive. Given time and isolation (no gene flow), population diversity will be reduced (P6). Based on their mosaic structure and high genetic distances, it seems that current H. pylori from Amerindians and Mestizos are in one of the intermediate states (P3′ or P5′). Arrows indicate introduction of new populations.
Similar articles
- Comparative study between Helicobacter pylori and host human genetics in the Dominican Republic.
Ono T, Cruz M, Jiménez Abreu JA, Nagashima H, Subsomwong P, Hosking C, Shiota S, Suzuki R, Yamaoka Y. Ono T, et al. BMC Evol Biol. 2019 Nov 1;19(1):197. doi: 10.1186/s12862-019-1526-9. BMC Evol Biol. 2019. PMID: 31675915 Free PMC article. - Helicobacter pylori genotyping from American indigenous groups shows novel Amerindian vacA and cagA alleles and Asian, African and European admixture.
Camorlinga-Ponce M, Perez-Perez G, Gonzalez-Valencia G, Mendoza I, Peñaloza-Espinosa R, Ramos I, Kersulyte D, Reyes-Leon A, Romo C, Granados J, Muñoz L, Berg DE, Torres J. Camorlinga-Ponce M, et al. PLoS One. 2011;6(11):e27212. doi: 10.1371/journal.pone.0027212. Epub 2011 Nov 3. PLoS One. 2011. PMID: 22073291 Free PMC article. - Phylogeographic evidence of cognate recognition site patterns and transformation efficiency differences in H. pylori: theory of strain dominance.
Maldonado-Contreras A, Mane SP, Zhang XS, Pericchi L, Alarcón T, Contreras M, Linz B, Blaser MJ, Domínguez-Bello MG. Maldonado-Contreras A, et al. BMC Microbiol. 2013 Sep 19;13:211. doi: 10.1186/1471-2180-13-211. BMC Microbiol. 2013. PMID: 24050390 Free PMC article. - The Story of Helicobacter pylori: Depicting Human Migrations from the Phylogeography.
Waskito LA, Yamaoka Y. Waskito LA, et al. Adv Exp Med Biol. 2019;1149:1-16. doi: 10.1007/5584_2019_356. Adv Exp Med Biol. 2019. PMID: 31016625 Review. - [Organization of the Helicobacter pylori genome].
Zawilak A, Zakrzewska-Czerwińska J. Zawilak A, et al. Postepy Hig Med Dosw. 2001;55(3):355-67. Postepy Hig Med Dosw. 2001. PMID: 11505637 Review. Polish.
Cited by
- Evolutionary History of the Helicobacter pylori Genome: Implications for Gastric Carcinogenesis.
Correa P, Piazuelo MB. Correa P, et al. Gut Liver. 2012 Jan;6(1):21-8. doi: 10.5009/gnl.2012.6.1.21. Epub 2012 Jan 12. Gut Liver. 2012. PMID: 22375167 Free PMC article. - Gastric cancer: an infectious disease.
Piazuelo MB, Epplein M, Correa P. Piazuelo MB, et al. Infect Dis Clin North Am. 2010 Dec;24(4):853-69, vii. doi: 10.1016/j.idc.2010.07.010. Infect Dis Clin North Am. 2010. PMID: 20937454 Free PMC article. Review. - Comparison of Small Gut and Whole Gut Microbiota of First-Degree Relatives With Adult Celiac Disease Patients and Controls.
Bodkhe R, Shetty SA, Dhotre DP, Verma AK, Bhatia K, Mishra A, Kaur G, Pande P, Bangarusamy DK, Santosh BP, Perumal RC, Ahuja V, Shouche YS, Makharia GK. Bodkhe R, et al. Front Microbiol. 2019 Feb 8;10:164. doi: 10.3389/fmicb.2019.00164. eCollection 2019. Front Microbiol. 2019. PMID: 30800106 Free PMC article. - Identification of New Helicobacter pylori Subpopulations in Native Americans and Mestizos From Peru.
Gutiérrez-Escobar AJ, Velapatiño B, Borda V, Rabkin CS, Tarazona-Santos E, Cabrera L, Cok J, Hooper CC, Jahuira-Arias H, Herrera P, Noureen M, Wang D, Romero-Gallo J, Tran B, Peek RM Jr, Berg DE, Gilman RH, Camargo MC. Gutiérrez-Escobar AJ, et al. Front Microbiol. 2020 Dec 14;11:601839. doi: 10.3389/fmicb.2020.601839. eCollection 2020. Front Microbiol. 2020. PMID: 33381095 Free PMC article. - Whole Genome Sequence and Phylogenetic Analysis Show Helicobacter pylori Strains from Latin America Have Followed a Unique Evolution Pathway.
Muñoz-Ramírez ZY, Mendez-Tenorio A, Kato I, Bravo MM, Rizzato C, Thorell K, Torres R, Aviles-Jimenez F, Camorlinga M, Canzian F, Torres J. Muñoz-Ramírez ZY, et al. Front Cell Infect Microbiol. 2017 Feb 28;7:50. doi: 10.3389/fcimb.2017.00050. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28293542 Free PMC article.
References
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. - PubMed
- Loureiro CL, Alonso R, Pacheco BA, Uzcategui MG, Villegas L, et al. High prevalence of GB virus C/hepatitis G virus genotype 3 among autochthonous Venezuelan populations. J Med Virol. 2002;68:357–362. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources