Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli - PubMed (original) (raw)
Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli
Natividad Ruiz. Proc Natl Acad Sci U S A. 2008.
Abstract
Peptidoglycan is a cell-wall glycopeptide polymer that protects bacteria from osmotic lysis. Whereas in gram-positive bacteria it also serves as scaffold for many virulence factors, in gram-negative bacteria, peptidoglycan is an anchor for the outer membrane. For years, we have known the enzymes required for the biosynthesis of peptidoglycan; what was missing was the flippase that translocates the lipid-anchored precursors across the cytoplasmic membrane before their polymerization into mature peptidoglycan. Using a reductionist bioinformatics approach, I have identified the essential inner-membrane protein MviN (renamed MurJ) as a likely candidate for the peptidoglycan flippase in Escherichia coli. Here, I present genetic and biochemical data that confirm the requirement of MurJ for peptidoglycan biosynthesis and that are in agreement with a role of MurJ as a flippase. Because of its essential nature, MurJ could serve as a target in the continuing search for antimicrobial compounds.
Conflict of interest statement
The author declares no conflict of interest.
Figures
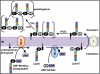
Fig. 1.
Biogenesis of peptidoglycan in E. coli. After being synthesized by MurABCDEF in the cytoplasm, UDP-MurNAc-pentapeptide is transferred to undecaprenyl phosphate by MraY, generating lipid I. Next, MurG synthesizes lipid II by adding GlcNAc (from UDP-GlcNAc) to lipid I. Lipid II is translocated across the IM by an unknown flippase. At the periplasmic side of the IM, glycosyltransferases (PGTs) assemble and elongate glycan chains, and transpeptidases (TPs) cross-link their peptide chains (dashed lines). DD-carboxypeptidases (CPs) remove terminal D-Ala from the stem pentapeptide in mature peptidoglycan.
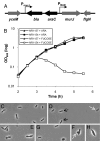
Fig. 2.
MurJ is essential. (A) Chromosomal organization of the MurJ-depletion strain. The PBAD promoter, araC and bla were inserted 14 bp upstream of the murJ start codon so that murJ expression is under the control of PBAD and decoupled from its native PmurJ promoter. (B) Growth (measured by OD600) of depletion strain NR1154 (MC4100 ara+ Δ_lysA_::kan murJ Ω(−14::bla araC PBAD) depends on the presence of arabinose in the medium. Addition of
d
-fucose, a nonmetabolizable analog of
l
-arabinose, causes cell lysis of NR1154 but not the MurJ+ parent strain NR1157 (MC4100 ara+ Δ_lysA_::kan). (C–H) Cell morphology (under magnification ×100 phase objective) of wild-type (NR1157, C) and MurJ-depletion (NR1154, D–H) strains grown in the presence of fucose. During MurJ depletion, whereas some cells appear wild type (black arrows), many are larger, irregularly shaped, and form large blebs (white arrows).
Similar articles
- The O-Antigen Flippase Wzk Can Substitute for MurJ in Peptidoglycan Synthesis in Helicobacter pylori and Escherichia coli.
Elhenawy W, Davis RM, Fero J, Salama NR, Felman MF, Ruiz N. Elhenawy W, et al. PLoS One. 2016 Aug 18;11(8):e0161587. doi: 10.1371/journal.pone.0161587. eCollection 2016. PLoS One. 2016. PMID: 27537185 Free PMC article. - Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis.
Sham LT, Butler EK, Lebar MD, Kahne D, Bernhardt TG, Ruiz N. Sham LT, et al. Science. 2014 Jul 11;345(6193):220-2. doi: 10.1126/science.1254522. Science. 2014. PMID: 25013077 Free PMC article. - The bacterial lipid II flippase MurJ functions by an alternating-access mechanism.
Kumar S, Rubino FA, Mendoza AG, Ruiz N. Kumar S, et al. J Biol Chem. 2019 Jan 18;294(3):981-990. doi: 10.1074/jbc.RA118.006099. Epub 2018 Nov 27. J Biol Chem. 2019. PMID: 30482840 Free PMC article. - Structure and Mechanism of the Lipid Flippase MurJ.
Kuk ACY, Hao A, Lee SY. Kuk ACY, et al. Annu Rev Biochem. 2022 Jun 21;91:705-729. doi: 10.1146/annurev-biochem-040320-105145. Epub 2022 Mar 23. Annu Rev Biochem. 2022. PMID: 35320686 Free PMC article. Review. - Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria.
Islam ST, Lam JS. Islam ST, et al. Environ Microbiol. 2013 Apr;15(4):1001-15. doi: 10.1111/j.1462-2920.2012.02890.x. Epub 2012 Sep 27. Environ Microbiol. 2013. PMID: 23016929 Review.
Cited by
- The Mycobacterial Cell Wall--Peptidoglycan and Arabinogalactan.
Alderwick LJ, Harrison J, Lloyd GS, Birch HL. Alderwick LJ, et al. Cold Spring Harb Perspect Med. 2015 Mar 27;5(8):a021113. doi: 10.1101/cshperspect.a021113. Cold Spring Harb Perspect Med. 2015. PMID: 25818664 Free PMC article. Review. - The O-Antigen Flippase Wzk Can Substitute for MurJ in Peptidoglycan Synthesis in Helicobacter pylori and Escherichia coli.
Elhenawy W, Davis RM, Fero J, Salama NR, Felman MF, Ruiz N. Elhenawy W, et al. PLoS One. 2016 Aug 18;11(8):e0161587. doi: 10.1371/journal.pone.0161587. eCollection 2016. PLoS One. 2016. PMID: 27537185 Free PMC article. - Bacillus subtilis homologs of MviN (MurJ), the putative Escherichia coli lipid II flippase, are not essential for growth.
Fay A, Dworkin J. Fay A, et al. J Bacteriol. 2009 Oct;191(19):6020-8. doi: 10.1128/JB.00605-09. Epub 2009 Aug 7. J Bacteriol. 2009. PMID: 19666716 Free PMC article. - Cutting edge: mutation of Francisella tularensis mviN leads to increased macrophage absent in melanoma 2 inflammasome activation and a loss of virulence.
Ulland TK, Buchan BW, Ketterer MR, Fernandes-Alnemri T, Meyerholz DK, Apicella MA, Alnemri ES, Jones BD, Nauseef WM, Sutterwala FS. Ulland TK, et al. J Immunol. 2010 Sep 1;185(5):2670-4. doi: 10.4049/jimmunol.1001610. Epub 2010 Aug 2. J Immunol. 2010. PMID: 20679532 Free PMC article. - What Is Motion? Recent Advances in the Study of Molecular Movement Patterns of the Peptidoglycan Synthesis Machines.
Lamanna MM, Maurelli AT. Lamanna MM, et al. J Bacteriol. 2022 Apr 19;204(4):e0059821. doi: 10.1128/JB.00598-21. Epub 2021 Dec 20. J Bacteriol. 2022. PMID: 34928180 Free PMC article. Review.
References
- Bouhss A, Trunkfield AE, Bugg TD, Mengin-Lecreulx D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev. 2008;32:208–233. - PubMed
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. - PubMed
- Dramsi S, Magnet S, Davison S, Arthur M. Covalent attachment of proteins to peptidoglycan. FEMS Microbiol Rev. 2008;32:307–320. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases