ARDB--Antibiotic Resistance Genes Database - PubMed (original) (raw)
. 2009 Jan;37(Database issue):D443-7.
doi: 10.1093/nar/gkn656. Epub 2008 Oct 2.
Affiliations
- PMID: 18832362
- PMCID: PMC2686595
- DOI: 10.1093/nar/gkn656
ARDB--Antibiotic Resistance Genes Database
Bo Liu et al. Nucleic Acids Res. 2009 Jan.
Abstract
The treatment of infections is increasingly compromised by the ability of bacteria to develop resistance to antibiotics through mutations or through the acquisition of resistance genes. Antibiotic resistance genes also have the potential to be used for bio-terror purposes through genetically modified organisms. In order to facilitate the identification and characterization of these genes, we have created a manually curated database--the Antibiotic Resistance Genes Database (ARDB)--unifying most of the publicly available information on antibiotic resistance. Each gene and resistance type is annotated with rich information, including resistance profile, mechanism of action, ontology, COG and CDD annotations, as well as external links to sequence and protein databases. Our database also supports sequence similarity searches and implements an initial version of a tool for characterizing common mutations that confer antibiotic resistance. The information we provide can be used as compendium of antibiotic resistance factors as well as to identify the resistance genes of newly sequenced genes, genomes, or metagenomes. Currently, ARDB contains resistance information for 13,293 genes, 377 types, 257 antibiotics, 632 genomes, 933 species and 124 genera. ARDB is available at http://ardb.cbcb.umd.edu/.
Figures
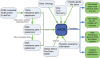
Figure 1.
ARDB curation data flow.
Figure 2.
Sample web pages from ARDB. (a) Front page, (b) resistance type, (c) blast result, (d) mutation annotation, (e) browse and (f) genome information.
Similar articles
- MvirDB--a microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications.
Zhou CE, Smith J, Lam M, Zemla A, Dyer MD, Slezak T. Zhou CE, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D391-4. doi: 10.1093/nar/gkl791. Epub 2006 Nov 7. Nucleic Acids Res. 2007. PMID: 17090593 Free PMC article. - u-CARE: user-friendly Comprehensive Antibiotic resistance Repository of Escherichia coli.
Saha SB, Uttam V, Verma V. Saha SB, et al. J Clin Pathol. 2015 Aug;68(8):648-51. doi: 10.1136/jclinpath-2015-202927. Epub 2015 May 2. J Clin Pathol. 2015. PMID: 25935546 - Cyanobacterial KnowledgeBase (CKB), a Compendium of Cyanobacterial Genomes and Proteomes.
Peter AP, Lakshmanan K, Mohandass S, Varadharaj S, Thilagar S, Abdul Kareem KA, Dharmar P, Gopalakrishnan S, Lakshmanan U. Peter AP, et al. PLoS One. 2015 Aug 25;10(8):e0136262. doi: 10.1371/journal.pone.0136262. eCollection 2015. PLoS One. 2015. PMID: 26305368 Free PMC article. - [Genetics and genomics for the study of bacterial resistance].
Garza-Ramos U, Silva-Sánchez J, Martínez-Romero E. Garza-Ramos U, et al. Salud Publica Mex. 2009;51 Suppl 3:S439-46. doi: 10.1590/s0036-36342009000900009. Salud Publica Mex. 2009. PMID: 20464217 Review. Spanish. - Rational identification of new antibacterial drug targets that are essential for viability using a genomics-based approach.
Chalker AF, Lunsford RD. Chalker AF, et al. Pharmacol Ther. 2002 Jul;95(1):1-20. doi: 10.1016/s0163-7258(02)00222-x. Pharmacol Ther. 2002. PMID: 12163125 Review.
Cited by
- Draft genome sequence of Escherichia coli LCT-EC106.
Li T, Pu F, Yang R, Fang X, Wang J, Guo Y, Chang D, Su L, Guo N, Jiang X, Zhao J, Liu C. Li T, et al. J Bacteriol. 2012 Aug;194(16):4443-4. doi: 10.1128/JB.00853-12. J Bacteriol. 2012. PMID: 22843582 Free PMC article. - Diminution of the gut resistome after a gut microbiota-targeted dietary intervention in obese children.
Wu G, Zhang C, Wang J, Zhang F, Wang R, Shen J, Wang L, Pang X, Zhang X, Zhao L, Zhang M. Wu G, et al. Sci Rep. 2016 Apr 5;6:24030. doi: 10.1038/srep24030. Sci Rep. 2016. PMID: 27044409 Free PMC article. - Draft genomes of four enterotoxigenic Escherichia coli (ETEC) clinical isolates from China and Bangladesh.
Liu F, Yang X, Wang Z, Nicklasson M, Qadri F, Yi Y, Zhu Y, Lv N, Li J, Zhang R, Guo H, Zhu B, Sjöling Å, Hu Y. Liu F, et al. Gut Pathog. 2015 Apr 8;7:10. doi: 10.1186/s13099-015-0059-z. eCollection 2015. Gut Pathog. 2015. PMID: 25932050 Free PMC article. - Translational metagenomics and the human resistome: confronting the menace of the new millennium.
Willmann M, Peter S. Willmann M, et al. J Mol Med (Berl). 2017 Jan;95(1):41-51. doi: 10.1007/s00109-016-1478-0. Epub 2016 Oct 20. J Mol Med (Berl). 2017. PMID: 27766372 Free PMC article. Review. - The species-level microbiota of healthy eyes revealed by the integration of metataxonomics with culturomics and genome analysis.
Dong K, Pu J, Yang J, Zhou G, Ji X, Kang Z, Li J, Yuan M, Ning X, Zhang Z, Ma X, Cheng Y, Li H, Ma Q, Li H, Zhao L, Lei W, Sun B, Xu J. Dong K, et al. Front Microbiol. 2022 Sep 2;13:950591. doi: 10.3389/fmicb.2022.950591. eCollection 2022. Front Microbiol. 2022. PMID: 36124162 Free PMC article.
References
- Bancroft EA. Antimicrobial resistance: it's not just for hospitals. JAMA. 2007;298:1803–1804. - PubMed
- Selvey LA, Whitby M, Johnson B. Nosocomial methicillin-resistant Staphylococcus aureus bacteremia: is it any worse than nosocomial methicillin-sensitive Staphylococcus aureus bacteremia? Infect. Control. Hosp. Epidemiol. 2000;21:645–648. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical