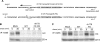Signal peptide-dependent inhibition of MHC class I heavy chain translation by rhesus cytomegalovirus - PubMed (original) (raw)
Signal peptide-dependent inhibition of MHC class I heavy chain translation by rhesus cytomegalovirus
Colin J Powers et al. PLoS Pathog. 2008.
Abstract
The US2-11 region of human and rhesus cytomegalovirus encodes a conserved family of glycoproteins that inhibit MHC-I assembly with viral peptides, thus preventing cytotoxic T cell recognition. Since HCMV lacking US2-11 is no longer able to block assembly and transport of MHC-I, we examined whether this is also observed for RhCMV lacking the corresponding region. Unexpectedly, recombinant RhCMV lacking US2-11 was still able to inhibit MHC-I expression in infected fibroblasts, suggesting the presence of an additional MHC-I evasion mechanism. Progressive deletion analysis of RhCMV-specific genomic regions revealed that MHC-I expression is fully restored upon additional deletion of rh178. The protein encoded by this RhCMV-specific open reading frame is anchored in the endoplasmic reticulum membrane. In the presence of rh178, RhCMV prevented MHC-I heavy chain (HC) expression, but did not inhibit mRNA transcription or association of HC mRNA with translating ribosomes. Proteasome inhibitors stabilized a HC degradation intermediate in the absence of rh178, but not in its presence, suggesting that rh178 prevents completion of HC translation. This interference was signal sequence-dependent since replacing the signal peptide with that of CD4 or murine HC rendered human HCs resistant to rh178. We have identified an inhibitor of antigen presentation encoded by rhesus cytomegalovirus unique in both its lack of homology to any other known protein and in its mechanism of action. By preventing signal sequence-dependent HC translocation, rh178 acts prior to US2, US3 and US11 which attack MHC-I proteins after protein synthesis is completed. Rh178 is the first viral protein known to interfere at this step of the MHC-I pathway, thus taking advantage of the conserved nature of HC leader peptides, and represents a new mechanism of translational interference.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
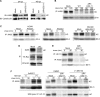
Figure 1. RhCMV inhibits HC expression in the absence of RhUS2-11.
All experiments were performed at 24 hours post infection at MOI = 3. A) Immunoblot analysis of MHC-I or calreticulin in Mock- or RhCMV-infected TRF lysates. B) IP of total MHC-I upon labeling with 35S-Met/Cys for the indicated time. (*) All IPs from WT and recombinant RhCMV- infected cells contain antibody-binding proteins around 55kDa (see Fig. S2) which likely correspond to the RhCMV homologues of the Fc-receptor UL119-118 of HCMV . Since these viral proteins are not involved in MHC-I inhibition they are not shown in most figures. C) Pulse-chase labeling of 10 min and immunoprecipitation of total MHC-I from Mock-infected, HCMV-infected THFs, or RhCMV-infected TRFs. D) Pulse-labeling of 60 min and IP of MHC-I, Tfn Rec (Transferrin receptor) or Vimentin from Mock-infected or RhCMV-infected TRFs. E) Pulse-chase labeling of 10 min and IP of total MHC-I or HC. Cells were labeled as in 1C, but lysed in SDS buffer prior to IP. F) Pulse-chase labeling and IP of RhCMV-infected TRFs treated with proteasome inhibitor. Where indicated TRFs were incubated with 50 µM MG132 or DMSO during 60-min of Met/Cys starvation, 10-min label, and 30-min chase. For control, TRFs were transduced with AdUS11 (MOI = 25), a recombinant adenovirus expressing HCMV US11, for 24 hours followed by NP40-lysis and IP with K455. Shown for RhCMV-infection is both NP-40 lysis (top panel) and SDS-lysis (bottom panel) prior to IP with the noted antibody.
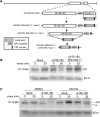
Figure 2. Deletion of Rh158–180 restores MHC-I expression during RhCMV infection.
A) Diagram of the step-wise construction of the ΔRhUS2-11 and Δ158–180,ΔRhUS2-11 viruses. Using the RhCMV BAC the RhUS2-11 region was replaced with a PCR-fragment containing a Kanamycin resistance (Kanr) cassette flanked by RhCMV homologous regions. The Kanr cassette was removed by arabinose-induced FLP recombinase prior to replacing the Rh158-180 region with Kanr. B) Pulse-chase labeling for 10 min of TRFs infected with WT or recombinant RhCMV followed by IP of total MHC-I. In C) 50 µM MG132 or DMSO was included as in Fig. 1F. (*) indicates a deglycosylated cytosolic degradation intermediate stabilized by MG132.
Figure 3. VIHCE is encoded by rh178.
A) Deletional mapping of VIHCE. Predicted open reading frames between Rh158–180 are shown as open white arrows. Solid black rectangles indicate the region of deletion. Pulse-chase labeling for 10 min with the indicated recombinant virus was performed as in Fig. 1C followed by IP with K455. Lack of VIHCE is readily apparent by the initial synthesis of HC (left lane) followed by US2-11-mediated destruction (right lane). B) Predicted ORFs and experimentally confirmed transcripts in the rh178 region. The red rectangle indicates the region essential for VIHCE function as determined by deletions in several independent recombinants. Large black arrows indicate positions of ORFs rh175–178 predicted by . Transcripts confirmed by RACE and cDNA PCR are shown below. C) Northern blot analysis of total RNA isolated from mock or WT RhCMV-infected TRFs at 24 hours post infection. ORF rh178 was used to generate 32P-dCTP labeled DNA probe. D) Complementary sequence of the RhCMV genome from 181921–182060bp. Underlined at 182058bp is the original predicted start codon for rh178 . Transcription actually begins at 182015bp as determined by 5′ RACE (see sequence chromatogram below genomic sequence). Shaded in gray is the first ATG codon of the transcript. Also noted is the splice donor site for rh178.4 which is spliced at 181944bp.
Figure 4. A frameshift mutation of rh178 restores HC expression.
A) Sequence of the rh178 frameshift control and frameshift recombinants. Shown is complementary genomic sequence, with transcripts running from right to left. In each recombinant a 20bp sequence in the 5′ UTR of rh178 (gray boxes) was replaced with 93bp from the recombination vector including the FRT recombination site. (*) indicates the single base insertion causing a frameshift. B) and C) HC expression in TRFs infected with control or frameshift viruses. Pulse-chase and IP was performed as in Fig. 1C.
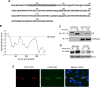
Figure 5. rh178 is a 212aa non-glycosylated ER localized protein that is sufficient to block HC synthesis.
A) Complete polypeptide sequence of rh178. Shaded in gray is the predicted signal anchor sequence. B) Hydrophobicity graph of rh178 (TopPred,
). TM refers to a predicted transmembrane domain cutoff value. C) Western blot of lysate from TRFs transduced with replication deficient adenovirus vectors AdTrans (expressing a tetracycline responsive transactivator) or AdTrans together with Ad178-HA (expressing HA-tagged rh178) for 48 hours. Lysate was treated without or with PNGase to remove N-linked sugars and blotted for MHC-I HC using the HC-10 antibody or for rh178-HA with an anti-HA antibody. D) HC expression in TRFs transduced with AdTrans or AdTrans with Ad178 (expressing wild type rh178) for 24 hours, followed by a 10-min pulse label and 30-min chase. HCs were recovered with K455 from NP40 lysates. E) Immunofluorescence analysis of TRFs 24 hours after transfection with HA-tagged rh178 together with FLAG-tagged K5 from KSHV. Primary antibodies were mouse anti-FLAG and rabbit anti-HA. Secondary antibodies were 594 Alexa Fluor conjugated goat anti-rabbit and 488 Alexa Fluor conjugated goat anti-mouse.
Figure 6. HC mRNA is present, intact, and associates with actively translating ribosomes during RhCMV infection.
A) Northern blot analysis of HC- or GAPDH-specific mRNA from total RNA isolated at 24 hours after Mock- or RhCMV-infection. The 32P-dCTP labeled probes were generated using rhesus-derived cDNAs for HC or GAPDH as templates. B) Polyribosome fractionation and northern blot analysis. TRFs were either mock infected or infected with wild-type (WT) RhCMV at MOI = 3 for 24 hours followed by isolation and fractionation of polysomes. Ethidium Bromide (EtBr) staining of a denaturing agarose gel shows the amount and ratio of 18S and 28S rRNA present in each fraction, indicating the presence of ribosomal subunits. Polysomes sediment to higher, denser fractions. Lower panels show northern blots of the gel using the HC and GAPDH-specific probes. C) Cells were infected as in B. However, after 24 hours, cells were incubated for 4 min with either DMSO or 100 µg/ml puromycin prior to polysome harvesting.
Figure 7. Efficient HC targeting by rh178 is signal-peptide dependent.
A) Rh178 inhibits expression of human HC. THFs were infected with the indicated virus at MOI = 3 for 24 hours, followed by a 10-min pulse-label, a chase of 30-min and IP with K455. B) UTR-independent inhibition of HLA-A3 expression by rh178. TRFs were electroporated with pEF1α containg the indicated HLA-A3 construct. After 24 hours, cells were either mock infected or infected with recombinant RhCMV (MOI = 3) containing VIHCE (+; ΔRhUS2-11) or lacking VIHCE (−; Δ178, ΔRhUS2-11). After an additional 24 hours, cells were labeled for 30-min, lysed in NP-40, and HLA-A3 was immunoprecipitated. C) Upper panel: Amino acid sequence of the signal peptides used in chimeric HLA-A3 HCs. Gray shading indicates identity with the HLA-A3 signal peptide. Lower panel: TRFs were electroporated with native HLA-A3 (A3) or the indicated SP-chimera (the HLA-A3 signal peptide was replaced with the H2-Kb or the CD4 signal peptide in Kb/A3 or CD4/A3, respectively) prior to infection with RhCMV, metabolic labeling and IP as in 7B. (*) indicates an uncharacterized HC-band that appears prominently in IPs from CD4/A3 transfectants and that could represent a deglycosylated or truncated HC. D) Quantitation of HLA-A3, total HC, or CD4 expression from 7C and 7E shown as a percent relative to HC or CD4 levels in the absence of VIHCE. Bands were quantitated using ImageQuant 5.1 software (Molecular Dynamics). E) TRFs were electroporated with native CD4 or CD4 containing the HLA-A3 SP (A3/CD4) and treated as in 7C. All experiments are representative of several replicates.
Similar articles
- The cytoplasmic domain of rhesus cytomegalovirus Rh178 interrupts translation of major histocompatibility class I leader peptide-containing proteins prior to translocation.
Richards R, Scholz I, Powers C, Skach WR, Früh K. Richards R, et al. J Virol. 2011 Sep;85(17):8766-76. doi: 10.1128/JVI.05021-11. Epub 2011 Jun 29. J Virol. 2011. PMID: 21715474 Free PMC article. - Rhesus cytomegalovirus contains functional homologues of US2, US3, US6, and US11.
Pande NT, Powers C, Ahn K, Früh K. Pande NT, et al. J Virol. 2005 May;79(9):5786-98. doi: 10.1128/JVI.79.9.5786-5798.2005. J Virol. 2005. PMID: 15827193 Free PMC article. - Inhibition of the MHC class II antigen presentation pathway by human cytomegalovirus.
Johnson DC, Hegde NR. Johnson DC, et al. Curr Top Microbiol Immunol. 2002;269:101-15. doi: 10.1007/978-3-642-59421-2_7. Curr Top Microbiol Immunol. 2002. PMID: 12224504 Review. - Inhibition of HLA-DR assembly, transport, and loading by human cytomegalovirus glycoprotein US3: a novel mechanism for evading major histocompatibility complex class II antigen presentation.
Hegde NR, Tomazin RA, Wisner TW, Dunn C, Boname JM, Lewinsohn DM, Johnson DC. Hegde NR, et al. J Virol. 2002 Nov;76(21):10929-41. doi: 10.1128/jvi.76.21.10929-10941.2002. J Virol. 2002. PMID: 12368336 Free PMC article. - Identifying the ERAD ubiquitin E3 ligases for viral and cellular targeting of MHC class I.
van den Boomen DJ, Lehner PJ. van den Boomen DJ, et al. Mol Immunol. 2015 Dec;68(2 Pt A):106-11. doi: 10.1016/j.molimm.2015.07.005. Epub 2015 Jul 22. Mol Immunol. 2015. PMID: 26210183 Free PMC article. Review.
Cited by
- MHC class I antigen presentation: learning from viral evasion strategies.
Hansen TH, Bouvier M. Hansen TH, et al. Nat Rev Immunol. 2009 Jul;9(7):503-13. doi: 10.1038/nri2575. Nat Rev Immunol. 2009. PMID: 19498380 Review. - Modulation of MHC-E transport by viral decoy ligands is required for RhCMV/SIV vaccine efficacy.
Verweij MC, Hansen SG, Iyer R, John N, Malouli D, Morrow D, Scholz I, Womack J, Abdulhaqq S, Gilbride RM, Hughes CM, Ventura AB, Ford JC, Selseth AN, Oswald K, Shoemaker R, Berkemeier B, Bosche WJ, Hull M, Shao J, Sacha JB, Axthelm MK, Edlefsen PT, Lifson JD, Picker LJ, Früh K. Verweij MC, et al. Science. 2021 Apr 30;372(6541):eabe9233. doi: 10.1126/science.abe9233. Epub 2021 Mar 25. Science. 2021. PMID: 33766941 Free PMC article. - Reevaluation of the coding potential and proteomic analysis of the BAC-derived rhesus cytomegalovirus strain 68-1.
Malouli D, Nakayasu ES, Viswanathan K, Camp DG 2nd, Chang WL, Barry PA, Smith RD, Früh K. Malouli D, et al. J Virol. 2012 Sep;86(17):8959-73. doi: 10.1128/JVI.01132-12. Epub 2012 Jun 20. J Virol. 2012. PMID: 22718821 Free PMC article. - The capacity of UL49.5 proteins to inhibit TAP is widely distributed among members of the genus Varicellovirus.
Verweij MC, Lipinska AD, Koppers-Lalic D, van Leeuwen WF, Cohen JI, Kinchington PR, Messaoudi I, Bienkowska-Szewczyk K, Ressing ME, Rijsewijk FA, Wiertz EJ. Verweij MC, et al. J Virol. 2011 Mar;85(5):2351-63. doi: 10.1128/JVI.01621-10. Epub 2010 Dec 15. J Virol. 2011. PMID: 21159875 Free PMC article. - The myeloid transcription factor GATA-2 regulates the viral UL144 gene during human cytomegalovirus latency in an isolate-specific manner.
Poole E, Walther A, Raven K, Benedict CA, Mason GM, Sinclair J. Poole E, et al. J Virol. 2013 Apr;87(8):4261-71. doi: 10.1128/JVI.03497-12. Epub 2013 Jan 30. J Virol. 2013. PMID: 23365437 Free PMC article.
References
- Pass RF. Cytomegalovirus. In: Knipe DavidM, PMH, Griffin DianeE, Lamb RobertA, Martin MalcolmA, Roizman Bernard, Straus StephenE., editors. Fields Virology. 4th ed. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2675–2705.
- Boppana SB, Rivera LB, Fowler KB, Mach M, Britt WJ. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N Engl J Med. 2001;344:1366–1371. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI059457/AI/NIAID NIH HHS/United States
- P51 RR000163-486829/RR/NCRR NIH HHS/United States
- R01 AI059457-01A2/AI/NIAID NIH HHS/United States
- P51 RR000163/RR/NCRR NIH HHS/United States
- RR 00163/RR/NCRR NIH HHS/United States
- P51 RR000163-496081/RR/NCRR NIH HHS/United States
- R01 AI059457-03/AI/NIAID NIH HHS/United States
- T32 AI 007472/AI/NIAID NIH HHS/United States
- T32 AI007472/AI/NIAID NIH HHS/United States
- K01 RR000163/RR/NCRR NIH HHS/United States
- R01 AI059457-02/AI/NIAID NIH HHS/United States
- R01 AI059457-04/AI/NIAID NIH HHS/United States
- R01 AI 0594457/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials