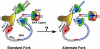Polymerase dynamics at the eukaryotic DNA replication fork - PubMed (original) (raw)
Review
Polymerase dynamics at the eukaryotic DNA replication fork
Peter M J Burgers. J Biol Chem. 2009.
Abstract
This review discusses recent insights in the roles of DNA polymerases (Pol) delta and epsilon in eukaryotic DNA replication. A growing body of evidence specifies Pol epsilon as the leading strand DNA polymerase and Pol delta as the lagging strand polymerase during undisturbed DNA replication. New evidence supporting this model comes from the use of polymerase mutants that show an asymmetric mutator phenotype for certain mispairs, allowing an unambiguous strand assignment for these enzymes. On the lagging strand, Pol delta corrects errors made by Pol alpha during Okazaki fragment initiation. During Okazaki fragment maturation, the extent of strand displacement synthesis by Pol delta determines whether maturation proceeds by the short or long flap processing pathway. In the more common short flap pathway, Pol delta coordinates with the flap endonuclease FEN1 to degrade initiator RNA, whereas in the long flap pathway, RNA removal is initiated by the Dna2 nuclease/helicase.
Figures
FIGURE 1.
Replication fork models. Left, model showing the primary replication functions of Pol δ and Pol ε; right, hypothetical replication fork formed by remodeling of the normal fork following replication stress or at specific chromosomal regions.
FIGURE 2.
Coordination between Pol δ and FEN1 in Okazaki fragment maturation. During Okazaki fragment maturation, Pol δ and FEN1 go through multiple cycles of displacement synthesis, molecular handoffs, and cutting (nick translation) until all initiator RNA (iRNA) has been degraded. During FEN1 dysfunction, idling maintains Pol δ at the nick position. Exo, exonuclease.
FIGURE 3.
Distribution between short and long flap removal pathways. The main pathway (thick arrows) involves limited strand displacement by Pol δ, followed by FEN1 cutting of the single nucleotide flap. This process is iterated until all initiator RNA (iRNA; red) is degraded (Nick Translation). Long flap formation (dashed arrows) results from excessive strand displacement synthesis. It is reduced by the exonuclease (Exo) activity of Pol δ (Idling) and promoted by the actions of Pol32 or Pif1. Dna2 cuts long flaps that are further degraded to precise nicks by FEN1 or the exonuclease activity of Pol δ (Long Flap Processing).
Similar articles
- Eukaryotic DNA Replication Fork.
Burgers PMJ, Kunkel TA. Burgers PMJ, et al. Annu Rev Biochem. 2017 Jun 20;86:417-438. doi: 10.1146/annurev-biochem-061516-044709. Epub 2017 Mar 1. Annu Rev Biochem. 2017. PMID: 28301743 Free PMC article. Review. - Dynamics of enzymatic interactions during short flap human Okazaki fragment processing by two forms of human DNA polymerase δ.
Lin SH, Wang X, Zhang S, Zhang Z, Lee EY, Lee MY. Lin SH, et al. DNA Repair (Amst). 2013 Nov;12(11):922-35. doi: 10.1016/j.dnarep.2013.08.008. Epub 2013 Sep 10. DNA Repair (Amst). 2013. PMID: 24035200 Free PMC article. - Reconstituted Okazaki fragment processing indicates two pathways of primer removal.
Rossi ML, Bambara RA. Rossi ML, et al. J Biol Chem. 2006 Sep 8;281(36):26051-61. doi: 10.1074/jbc.M604805200. Epub 2006 Jul 11. J Biol Chem. 2006. PMID: 16837458 - Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal.
Rossi ML, Pike JE, Wang W, Burgers PMJ, Campbell JL, Bambara RA. Rossi ML, et al. J Biol Chem. 2008 Oct 10;283(41):27483-27493. doi: 10.1074/jbc.M804550200. Epub 2008 Aug 9. J Biol Chem. 2008. PMID: 18689797 Free PMC article. - Mechanism of Lagging-Strand DNA Replication in Eukaryotes.
Stodola JL, Burgers PM. Stodola JL, et al. Adv Exp Med Biol. 2017;1042:117-133. doi: 10.1007/978-981-10-6955-0_6. Adv Exp Med Biol. 2017. PMID: 29357056 Review.
Cited by
- Replicative DNA polymerases.
Johansson E, Dixon N. Johansson E, et al. Cold Spring Harb Perspect Biol. 2013 Jun 1;5(6):a012799. doi: 10.1101/cshperspect.a012799. Cold Spring Harb Perspect Biol. 2013. PMID: 23732474 Free PMC article. Review. - DNA polymerase swapping in Caudoviricetes bacteriophages.
Yutin N, Tolstoy I, Mutz P, Wolf YI, Krupovic M, Koonin EV. Yutin N, et al. Virol J. 2024 Aug 26;21(1):200. doi: 10.1186/s12985-024-02482-z. Virol J. 2024. PMID: 39187833 Free PMC article. - Evolution of replication machines.
Yao NY, O'Donnell ME. Yao NY, et al. Crit Rev Biochem Mol Biol. 2016 May-Jun;51(3):135-49. doi: 10.3109/10409238.2015.1125845. Epub 2015 Dec 20. Crit Rev Biochem Mol Biol. 2016. PMID: 27160337 Free PMC article. Review. - Evolution of Eukaryotic DNA Polymerases via Interaction Between Cells and Large DNA Viruses.
Takemura M, Yokobori S, Ogata H. Takemura M, et al. J Mol Evol. 2015 Aug;81(1-2):24-33. doi: 10.1007/s00239-015-9690-z. Epub 2015 Jul 16. J Mol Evol. 2015. PMID: 26177821 - Pre-steady state kinetic studies of the fidelity of nucleotide incorporation by yeast DNA polymerase delta.
Dieckman LM, Johnson RE, Prakash S, Washington MT. Dieckman LM, et al. Biochemistry. 2010 Aug 31;49(34):7344-50. doi: 10.1021/bi100556m. Biochemistry. 2010. PMID: 20666462 Free PMC article.
References
- Waga, S., and Stillman, B. (1998) Annu. Rev. Biochem. 67 721–751 - PubMed
- Garg, P., and Burgers, P. M. (2005) Crit. Rev. Biochem. Mol. Biol. 40 115–128 - PubMed
- Johnson, A., and O'Donnell, M. (2005) Annu. Rev. Biochem. 74 283–315 - PubMed
- Morrison, A., Araki, H., Clark, A. B., Hamatake, R. K., and Sugino, A. (1990) Cell 62 1143–1151 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous