The Bacillus anthracis SleL (YaaH) protein is an N-acetylglucosaminidase involved in spore cortex depolymerization - PubMed (original) (raw)
The Bacillus anthracis SleL (YaaH) protein is an N-acetylglucosaminidase involved in spore cortex depolymerization
Emily A Lambert et al. J Bacteriol. 2008 Dec.
Abstract
Bacillus anthracis spores, the infectious agents of anthrax, are notoriously difficult to remove from contaminated areas because they are resistant to many eradication methods. These resistance properties are due to the spore's dehydration and dormancy and to the multiple protective layers surrounding the spore core, one of which is the cortex. In order for B. anthracis spores to germinate and resume growth, the cortex peptidoglycan must be depolymerized. This study reports on analyses of sleL (yaaH), which encodes a cortex-lytic enzyme. The inactivation of sleL does not affect vegetative growth, spore viability, or the initial stages of germination, including dipicolinic acid release. However, mutant spores exhibit a slight delay in the loss of optical density compared to that of wild-type spores. Mutants also retain more diaminopimelic acid and N-acetylmuramic acid during germination than wild-type spores, suggesting that the cortex peptidoglycan is not being hydrolyzed as rapidly. This finding is supported by high-pressure liquid chromatography analysis of the peptidoglycan structure used to confirm that SleL acts as an N-acetylglucosaminidase. When sleL is inactivated, the cortex peptidoglycan is not depolymerized into small muropeptides but instead is retained within the spore as large fragments. In the absence of the sleL-encoded N-acetylglucosaminidase, other cortex-lytic enzymes break down the cortex peptidoglycan sufficiently to allow rapid germination and outgrowth.
Figures
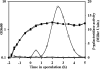
FIG. 1.
Sporulation-specific expression of sleL. Strains were incubated in modified G medium at 39°C for 7 h, during which samples were collected for β-galactosidase assays. Growth (filled symbols) and β-galactosidase activity (open symbols) were assayed for wild-type (circles) and DPBa27 (sleL-lacZ::pDPV350) (squares) B. anthracis. The graph is representative of three independent experiments.
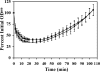
FIG. 2.
Germination of sleL spores is slightly delayed. Heat-activated spores were germinated in BHI medium at 39°C. Germination and outgrowth of wild-type (•) and DPBa27 (sleL-lacZ::pDPV350) (▪) B. anthracis spores were followed as changes in OD600 value over time. Error bars represent 1 standard deviation of the mean of three independent assays. The difference in the loss of OD600 between the two strains is significant (P ≤ 0.0098) from 10 to 35 min after germination initiation.
FIG. 3.
Release of DPA and NAM during germination. Heat-activated spores were germinated with
l
-alanine and inosine in buffer at 37°C for 10 min, and samples were removed and centrifuged for assay of exudate and spore pellet contents. The percentages of DPA and NAM released from B. anthracis spores of the wild type (black bars), the Δ_sleL_ mutant (DPBa35; white bars), the Δ_sleL_ mutant with pBKJ236 vector control (DPBa36; light gray bars), and the Δ_sleL_ mutant with pDPV352 complementing vector (DPBa37; dark gray bars) are shown. Error bars represent 1 standard deviation of the mean of three independent experiments. Asterisks indicate a statistically significant difference (P ≤ 0.0001) compared to wild-type B. anthracis. A cross indicates a statistically significant difference (P ≤ 0.007) compared to either the wild-type or DPBa35.
FIG. 4.
HPLC analysis of wild-type and Δ_sleL_ mutant B. anthracis muropeptides. Heat-activated spores were germinated for 5 min with
l
-alanine and inosine in buffer at 37°C. Samples were removed and centrifuged for analysis of PG in the exudate and spore pellet. PG was collected, digested, and reduced, and muropeptides were separated using a methanol gradient as previously described (11). Muropeptides were derived from dormant spores (A), from the exudates of germinating spores (B to E), from the exudates of germinating spores digested with mutanolysin (F and G), and from germinating spore pellets (H and I). Samples were from spores of the wild type (A, B, F, and H), the Δ_sleL_ mutant (DPBa35) (C, G, and I), the Δ_sleL_ mutant with pBKJ236 vector control (DPBa36) (D), and the Δ_sleL_ mutant with pDPV352 complementing vector (DPBa37) (E). Peaks G3, G10u, and G12 are clearly visible upon the magnification of panel E but are not marked for clarity. Peaks are labeled as in reference and Table 2, except the first “a” in the germination-specific peak names is omitted for space considerations. The identities of peaks aG7a and aG7b are unpublished data. Ino indicates the added germinant inosine. Large unlabeled peaks eluting between 10 and 30 min for exudate samples are other small molecules released from germinating spores that were analyzed and found not to contain peptidoglycan components. Peaks labeled b are buffer components.
Similar articles
- In vitro and in vivo analyses of the Bacillus anthracis spore cortex lytic protein SleL.
Lambert EA, Sherry N, Popham DL. Lambert EA, et al. Microbiology (Reading). 2012 May;158(Pt 5):1359-1368. doi: 10.1099/mic.0.056630-0. Epub 2012 Feb 16. Microbiology (Reading). 2012. PMID: 22343356 Free PMC article. - Contributions of four cortex lytic enzymes to germination of Bacillus anthracis spores.
Heffron JD, Lambert EA, Sherry N, Popham DL. Heffron JD, et al. J Bacteriol. 2010 Feb;192(3):763-70. doi: 10.1128/JB.01380-09. Epub 2009 Dec 4. J Bacteriol. 2010. PMID: 19966006 Free PMC article. - Cortex peptidoglycan lytic activity in germinating Bacillus anthracis spores.
Dowd MM, Orsburn B, Popham DL. Dowd MM, et al. J Bacteriol. 2008 Jul;190(13):4541-8. doi: 10.1128/JB.00249-08. Epub 2008 May 2. J Bacteriol. 2008. PMID: 18456807 Free PMC article. - Spore Peptidoglycan.
Popham DL, Bernhards CB. Popham DL, et al. Microbiol Spectr. 2015 Dec;3(6). doi: 10.1128/microbiolspec.TBS-0005-2012. Microbiol Spectr. 2015. PMID: 27337277 Review. - The role of peptidoglycan structure and structural dynamics during endospore dormancy and germination.
Atrih A, Foster SJ. Atrih A, et al. Antonie Van Leeuwenhoek. 1999 May;75(4):299-307. doi: 10.1023/a:1001800507443. Antonie Van Leeuwenhoek. 1999. PMID: 10510717 Review.
Cited by
- Identification of Universally Applicable and Species-Specific Marker Peptides for Bacillus anthracis.
Witt N, Galante D, Andreotti S, Abdel Glil M, Fasanella A, Meierhofer D, Tomaso H. Witt N, et al. Life (Basel). 2022 Oct 6;12(10):1549. doi: 10.3390/life12101549. Life (Basel). 2022. PMID: 36294983 Free PMC article. - Detecting Cortex Fragments During Bacterial Spore Germination.
Francis MB, Sorg JA. Francis MB, et al. J Vis Exp. 2016 Jun 25;(112):54146. doi: 10.3791/54146. J Vis Exp. 2016. PMID: 27403726 Free PMC article. - In vitro and in vivo analyses of the Bacillus anthracis spore cortex lytic protein SleL.
Lambert EA, Sherry N, Popham DL. Lambert EA, et al. Microbiology (Reading). 2012 May;158(Pt 5):1359-1368. doi: 10.1099/mic.0.056630-0. Epub 2012 Feb 16. Microbiology (Reading). 2012. PMID: 22343356 Free PMC article. - SpoVID functions as a non-competitive hub that connects the modules for assembly of the inner and outer spore coat layers in Bacillus subtilis.
Nunes F, Fernandes C, Freitas C, Marini E, Serrano M, Moran CP Jr, Eichenberger P, Henriques AO. Nunes F, et al. Mol Microbiol. 2018 Nov;110(4):576-595. doi: 10.1111/mmi.14116. Epub 2018 Oct 18. Mol Microbiol. 2018. PMID: 30168214 Free PMC article. - Contributions of four cortex lytic enzymes to germination of Bacillus anthracis spores.
Heffron JD, Lambert EA, Sherry N, Popham DL. Heffron JD, et al. J Bacteriol. 2010 Feb;192(3):763-70. doi: 10.1128/JB.01380-09. Epub 2009 Dec 4. J Bacteriol. 2010. PMID: 19966006 Free PMC article.
References
- Atrih, A., G. Bacher, R. Korner, G. Allmaier, and S. J. Foster. 1999. Structural analysis of Bacillus megaterium KM spore peptidoglycan and its dynamics during germination. Microbiology 1451033-1041. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources