Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS - PubMed (original) (raw)
Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS
Min Wu et al. Mol Cell Biol. 2008 Dec.
Abstract
In yeast, the macromolecular complex Set1/COMPASS is capable of methylating H3K4, a posttranslational modification associated with actively transcribed genes. There is only one Set1 in yeast; yet in mammalian cells there are multiple H3K4 methylases, including Set1A/B, forming human COMPASS complexes, and MLL1-4, forming human COMPASS-like complexes. We have shown that Wdr82, which associates with chromatin in a histone H2B ubiquitination-dependent manner, is a specific component of Set1 complexes but not that of MLL1-4 complexes. RNA interference-mediated knockdown of Wdr82 results in a reduction in the H3K4 trimethylation levels, although these cells still possess active MLL complexes. Comprehensive in vitro enzymatic studies with Set1 and MLL complexes demonstrated that the Set1 complex is a more robust H3K4 trimethylase in vitro than the MLL complexes. Given our in vivo and in vitro observations, it appears that the human Set1 complex plays a more widespread role in H3K4 trimethylation than do the MLL complexes in mammalian cells.
Figures
FIG. 1.
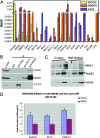
Wdr82 is a component of the Set1 complex that associates with chromatin in an H2B monoubiquitination-dependent manner. (A) Mammalian H3K4 trimethylase complexes were affinity purified through Flag-Wdr82, Flag-RBBP5, or Flag-Ash2. Relative protein levels were estimated by their dNSAF values (calculated based on unique spectral counts and shared spectral counts distributed among isoforms). dNSAF values were averaged across three (Wdr82) or two (RBBP5 and Ash2) independent runs. (B) Extracts from the Flag-tagged PTIP HeLa stable cells were immunoprecipitated with rabbit immunoglobulin G (IgG), anit-Flag, anti-MLL2, anti-Rbbp5 antibodies, followed by Western blotting with anti-Rbbp5, anti-Wdr82, and anti-Flag antibodies. (C) Human Bre1 (RNF20/40) were co-knocked down in Flag-Wdr82 stable cell lines via RNAi. Increasing amounts of cell extract were analyzed by Western analysis. (D) ChIP assays were carried out with anti-Flag antibody with the same samples as in panel C. The 3′-untranslated region of hemoglobin gene was used as a nonexpressing control gene internal control. The recruitment of Flag-Wdr82 to GAPDH, PP1A, and PABPC-1 promoters was decreased in the H2B ubiquitination-deficient cells. The asterisk in panel C indicates the presence of the nonspecific bands in the Western analysis.
FIG. 2.
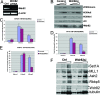
Reduction of Wdr82 levels results in the loss of H3K4 trimethylation and Set1A. (A) RNAi knockdown of Wdr82 in HeLa cells was confirmed by reverse transcription-PCR. (B) RNAi knockdown of Wdr82 in HeLa cells results in a reduction in H3K4 trimethylation levels but not a reduction in H3K4 mono- and dimethylation. WDR82 mRNA was knocked down using RNAi. Cell extracts from mock RNAi and WDR82 RNAi were tested by Western analysis with antibodies specific to mono-, di-, and trimethylated H3K4. (C to E) Wdr82 mRNA levels were knocked down in HeLa cells with siRNA, and CHIP assays were performed with H3K4 mono-, di-, and trimethylation antibodies on the promoters of GAPDH (C), PP1A (D), and PABPC1 (E) genes. The H3K4 trimethylation of all three genes decreased approximately two- to threefold, while the dimethylation was relatively unchanged for GAPDH and PP1A, and no changes were observed for PABPC1. (F) RNAi knockdown of Wdr82 results in the reduction in Set1A levels with no effect on MLL1, Ash2, or Rbbp5 levels. Reductions in the levels of WDR82, result in destabilization of Set1 complexes. The asterisks in panels B and F indicate the presence of nonspecific bands in the Western analysis.
FIG. 3.
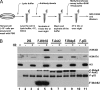
In vitro H3K4 trimethylase activities. (A) Schematic flowchart of the in vitro histone H3K4 methyltransferase assay with either human MLL- or Set1-containing complexes. (B) In vitro HMTase assays were performed with the complexes purified by anti-Flag antibody. HMTase was immunoprecipitated from either Flag-Wdr82, Flag-Ash2, Flag-Rbbp5, or Flag-p53 cell lines and tested for H3K4 di- and trimethylase activities.
FIG. 4.
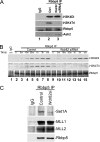
Role of Wdr82 in H3K4 trimethylation in vitro. (A) The loss of Wdr82 via RNAi results in the purification of HMTase complexes with low H3K4 trimethylase activity. Wdr82 was knocked down in HeLa cells bearing Flag-Rbbp5, which is a common subunit of the MLLs and Set1-containing complexes. After a Flag immunoprecipitation, the effect of Wdr82 reduction on H3K4 trimethylation activities was tested. In the absence of Wdr82, the H3K4 trimethylation activities of Rbbp5-containing complexes were reduced. (B) Kinetics of H3K4 di- and trimethylation in the presence or absence of Wdr82. HMTase assays with Rbbp5 purified complexes as in panel A were performed at different time points. The dimethylation activity was retarded in the Wdr82 knockdown samples, but both the control and the knockdown samples reached the similar maximum levels after 3 h. However, the trimethylation activities were decreased substantially after Wdr82 knockdown. (C) To demonstrate the effect of Wdr82 RNAi on Set1, MLL, and MLL2, and their complex stabilities, Rbbp5 purified complexes from Wdr82 knockdown were tested by Western analysis with antibodies to Set1, MLL1, and MLL2 and a shared member of their complexes RBbp5. This panel demonstrates a decrease in Set1A levels but not in MLL1 and MLL2 complexes as a result of Wdr82 RNAi.
FIG. 5.
Comparative analyses of the HMTase activities of the Set1 and MLL complexes in vivo and in vitro. (A) The levels of Set1A and Set1B were knocked down in HeLa cells by siRNA either alone or together. H3K4 mono-, di-, and trimethylation was tested in the resulting cell extracts. Compared to the control, H3K4 trimethylation decreased in all of the knockdown samples, with most in double knockdown, while the dimethylation levels did not change. (B) The total histone H3K4 methylation status was tested by Western blot analysis in MLL1+/+ and MLL1−/− MEF cells. The total levels of mono-, di-, and trimethylation were unchanged in the MLL1 knockout cell line. (C) MLL3/4 trimethylase activity in comparison to the Set1 complexes. Flag immunoprecipitation was performed in Flag-PTIP and Flag-Wdr82 stable cell lines to purify the MLL3/4 and Set1 complexes, respectively. In vitro HMTase assays were performed with each purified complex. The level of Rbbp5 was used for normalization of the purified HMTase complexes. With the same amount of Rbbp5, the Set1 complexes demonstrated a much more robust H3K4 trimethylase activity than did the MLL3/4 complexes. (D) MLL2 trimethylase activity in comparison to Set1 complexes. After their purification, the MLL2 and Set1 complexes were tested for HMT activities. With the same amount of Rbbp5, the Set1 complexes demonstrate a much more robust H3K4 trimethylase activity than did the MLL2 complex.
Similar articles
- Regulation of H3K4 trimethylation via Cps40 (Spp1) of COMPASS is monoubiquitination independent: implication for a Phe/Tyr switch by the catalytic domain of Set1.
Takahashi YH, Lee JS, Swanson SK, Saraf A, Florens L, Washburn MP, Trievel RC, Shilatifard A. Takahashi YH, et al. Mol Cell Biol. 2009 Jul;29(13):3478-86. doi: 10.1128/MCB.00013-09. Epub 2009 Apr 27. Mol Cell Biol. 2009. PMID: 19398585 Free PMC article. - Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II.
Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, Ge K, Krumlauf R, Shilatifard A. Wang P, et al. Mol Cell Biol. 2009 Nov;29(22):6074-85. doi: 10.1128/MCB.00924-09. Epub 2009 Aug 24. Mol Cell Biol. 2009. PMID: 19703992 Free PMC article. - Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation.
Vitaliano-Prunier A, Menant A, Hobeika M, Géli V, Gwizdek C, Dargemont C. Vitaliano-Prunier A, et al. Nat Cell Biol. 2008 Nov;10(11):1365-71. doi: 10.1038/ncb1796. Epub 2008 Oct 12. Nat Cell Biol. 2008. PMID: 18849979 - WRAD: enabler of the SET1-family of H3K4 methyltransferases.
Ernst P, Vakoc CR. Ernst P, et al. Brief Funct Genomics. 2012 May;11(3):217-26. doi: 10.1093/bfgp/els017. Epub 2012 May 30. Brief Funct Genomics. 2012. PMID: 22652693 Free PMC article. Review. - The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis.
Shilatifard A. Shilatifard A. Annu Rev Biochem. 2012;81:65-95. doi: 10.1146/annurev-biochem-051710-134100. Annu Rev Biochem. 2012. PMID: 22663077 Free PMC article. Review.
Cited by
- Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes.
van Nuland R, Smits AH, Pallaki P, Jansen PW, Vermeulen M, Timmers HT. van Nuland R, et al. Mol Cell Biol. 2013 May;33(10):2067-77. doi: 10.1128/MCB.01742-12. Epub 2013 Mar 18. Mol Cell Biol. 2013. PMID: 23508102 Free PMC article. - Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory.
D'Urso A, Takahashi YH, Xiong B, Marone J, Coukos R, Randise-Hinchliff C, Wang JP, Shilatifard A, Brickner JH. D'Urso A, et al. Elife. 2016 Jun 23;5:e16691. doi: 10.7554/eLife.16691. Elife. 2016. PMID: 27336723 Free PMC article. - Coordinated regulation of cellular identity-associated H3K4me3 breadth by the COMPASS family.
Sze CC, Ozark PA, Cao K, Ugarenko M, Das S, Wang L, Marshall SA, Rendleman EJ, Ryan CA, Zha D, Douillet D, Chen FX, Shilatifard A. Sze CC, et al. Sci Adv. 2020 Jun 24;6(26):eaaz4764. doi: 10.1126/sciadv.aaz4764. eCollection 2020 Jun. Sci Adv. 2020. PMID: 32637595 Free PMC article. - The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Zhang P, Lee H, Brunzelle JS, Couture JF. Zhang P, et al. Nucleic Acids Res. 2012 May;40(9):4237-46. doi: 10.1093/nar/gkr1235. Epub 2012 Jan 20. Nucleic Acids Res. 2012. PMID: 22266653 Free PMC article. - Modification of enhancer chromatin: what, how, and why?
Calo E, Wysocka J. Calo E, et al. Mol Cell. 2013 Mar 7;49(5):825-37. doi: 10.1016/j.molcel.2013.01.038. Mol Cell. 2013. PMID: 23473601 Free PMC article. Review.
References
- Bernstein, B. E., A. Meissner, and E. S. Lander. 2007. The mammalian epigenome. Cell 128669-681. - PubMed
- Dou, Y., T. A. Milne, A. J. Ruthenburg, S. Lee, J. W. Lee, G. L. Verdine, C. D. Allis, and R. G. Roeder. 2006. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13713-719. - PubMed
- Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 27728368-28371. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases