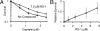A chemical compound that stimulates the human homologous recombination protein RAD51 - PubMed (original) (raw)
A chemical compound that stimulates the human homologous recombination protein RAD51
Krishanthi Jayathilaka et al. Proc Natl Acad Sci U S A. 2008.
Abstract
RAD51 and other members of the RecA family of strand exchange proteins assemble on ssDNA to form presynaptic filaments, which carry out the central steps of homologous recombination. A microplate-based assay was developed for high-throughput measurement of hRAD51 filament formation on ssDNA. With this method, a 10,000 compound library was screened, leading to the identification of a small molecule (RS-1) that enhances hRAD51 binding in a wide range of biochemical conditions. Salt titration experiments showed that RS-1 can enhance filament stability. Ultrastructural analysis of filaments formed on ssDNA showed that RS-1 can increase both protein-DNA complex lengths and the pitch of helical filament turns. RS-1 stimulated hRAD51-mediated homologous strand assimilation (D-loop) activity by at least 5- to 11-fold, depending on the condition. This D-loop stimulation occurred even in the presence of Ca(2+) or adenylyl-imidodiphosphate, indicating that the mechanism of stimulation was distinct from that conferred by Ca(2+) and/or inhibition of ATPase. No D-loop activity was observed in the absence of a nucleotide triphosphate cofactor, indicating that the compound does not substitute for this requirement. These results indicate that RS-1 enhances the homologous recombination activity of hRAD51 by promoting the formation of active presynaptic filaments. Cell survival assays in normal neonatal human dermal fibroblasts demonstrated that RS-1 promotes a dose-dependent resistance to the cross-linking chemotherapeutic drug cisplatin. Given that RAD51-dependent recombination is a major determinant of cisplatin resistance, RS-1 seems to function in vivo to stimulate homologous recombination repair proficiency. RS-1 has many potential applications in both research and medical settings.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
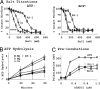
Effects of RS-1 on DNA binding. Various concentrations of DNA strand exchange proteins were incubated with 10 nM polydT containing a 5′ OG tag. Reactions were performed in Buffer B in the absence (diamonds) or presence of 20 μM RS-1 (squares). (A) hRAD51 in ATP and 10 mM MgCl2. (B) hRAD51 in ATP and 1 mM CaCl2 + 10 mM MgCl2. (C) Quantification of DNA binding by DNA strand exchange proteins under various conditions. NS, no statistically significant reduction.
Fig. 2.
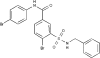
The chemical structure of RS-1 (3-[(benzylamino)sulfonyl]-4-bromo-N-(4-bromophenyl)benzamide).
Fig. 3.
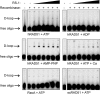
RS-1 stimulates hRAD51 via a novel mechanism. (A) RS-1, but not CaCl2, stabilizes hRAD51 nucleoprotein filaments formed in the presence of ADP. hRAD51 (500 nM) was incubated for 30 min with 10 nM OG-tagged polydT in Buffer B containing various combinations of 2 mM nucleotide cofactor, 1 mM CaCl2 (in addition to 10 mM MgCl2), and/or 20 μM RS-1, as indicated. Protein–DNA complexes were subsequently challenged by the addition of various concentrations of NaCl. (B) RS-1 does not inhibit hRAD51's ATPase activity. DNA-dependent ATP hydrolysis (shown by solid lines) was measured after incubation of 2 μM hRAD51 with 6 μM M13 ssDNA under conditions described in
SI Methods
. Buffers contained either 10 mM MgCl2, 5 mM CaCl2, or 10 mM MgCl2 + 20 μM RS-1, as indicated. Dotted lines show repeat experiments performed without DNA. (C) ATP but not RS-1 allows the DNA binding activity of hRAD51 to tolerate pretreatment with MgCl2. hRAD51 was preincubated at for 30 min at 37°C in Buffer B in combination with either 2 mM ATP, 20 μM RS-1 with no nucleotide cofactor, or neither, as indicated. OG-tagged polydT (10 nM) and 2 mM ATP were subsequently added, and the mixtures were incubated for an additional 30 min before FP was measured.
Fig. 4.
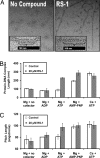
RS-1 affects the average length and helical pitch of hRAD51 protein–DNA complexes. Electron microscopy images of hRAD51 filaments formed on ssDNA were obtained at a magnification of ×25,000. Fifty randomly selected filaments were analyzed for each reaction condition. (A) RS-1 alters protein–DNA complex length and appearance. Representative images for filaments formed in ATP and Mg2+ plus the presence (Right) or absence (Left) of 20 μM RS-1. Insets at higher magnification demonstrate differences in length (brackets indicate 10 striations and bars denotes 100 nm). (B) Forty to fifty unselected protein–DNA complexes were measured. Cofactor requirements for RS-1 stimulation are displayed, and error bars represent SD. (C) RS-1 has a similar effect as AMP-PNP or Ca2+ on filament pitch (magnification was ×39,000).
Fig. 5.
RS-1 stimulates strand assimilation activity of hRAD51. Various DNA strand exchange proteins were allowed to form joint molecules (“D-loops”) in the presence of RS-1 (concentration range: 0, 1, 5, 10, 15, 20, and 25 μM). Reactions were performed as described in Methods, and phosphorimages of agarose gels are displayed for various conditions, as indicated. The positions of free oligo and oligo associated with D-loops are indicated.
Fig. 6.
RS-1 promotes resistance of human cells to cross-linking chemotherapy. Survival analyses were performed on neonatal human dermal fibroblasts. Cells were incubated for 24 h in media containing varying concentrations of cisplatin in the presence or absence of RS-1. Drugs were then removed, and cells were allowed to grow in complete media for an additional 6 days. Survival was measured with an sulforhodamine B method, as described in Methods. (A) Cisplatin titration shows that RS-1 at 7.5 μM promotes a resistance to cisplatin. (B) RS-1 titration at 1.2 μM cisplatin shows a concentration-dependent RS-1 effect.
Similar articles
- Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity.
Bugreev DV, Mazin AV. Bugreev DV, et al. Proc Natl Acad Sci U S A. 2004 Jul 6;101(27):9988-93. doi: 10.1073/pnas.0402105101. Epub 2004 Jun 28. Proc Natl Acad Sci U S A. 2004. PMID: 15226506 Free PMC article. - The human Rad51 protein: polarity of strand transfer and stimulation by hRP-A.
Baumann P, West SC. Baumann P, et al. EMBO J. 1997 Sep 1;16(17):5198-206. doi: 10.1093/emboj/16.17.5198. EMBO J. 1997. PMID: 9311980 Free PMC article. - Without Binding ATP, Human Rad51 Does Not Form Helical Filaments on ssDNA.
Schay G, Borka B, Kernya L, Bulyáki É, Kardos J, Fekete M, Fidy J. Schay G, et al. J Phys Chem B. 2016 Mar 10;120(9):2165-78. doi: 10.1021/acs.jpcb.5b12220. Epub 2016 Mar 1. J Phys Chem B. 2016. PMID: 26890079 - Assembly of RecA-like recombinases: distinct roles for mediator proteins in mitosis and meiosis.
Gasior SL, Olivares H, Ear U, Hari DM, Weichselbaum R, Bishop DK. Gasior SL, et al. Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8411-8. doi: 10.1073/pnas.121046198. Proc Natl Acad Sci U S A. 2001. PMID: 11459983 Free PMC article. Review. - Targeting the homologous recombination pathway by small molecule modulators.
Huang F, Mazin AV. Huang F, et al. Bioorg Med Chem Lett. 2014 Jul 15;24(14):3006-13. doi: 10.1016/j.bmcl.2014.04.088. Epub 2014 May 6. Bioorg Med Chem Lett. 2014. PMID: 24856061 Free PMC article. Review.
Cited by
- CRISPR-Cas9-mediated homology-directed repair for precise gene editing.
Liao H, Wu J, VanDusen NJ, Li Y, Zheng Y. Liao H, et al. Mol Ther Nucleic Acids. 2024 Sep 26;35(4):102344. doi: 10.1016/j.omtn.2024.102344. eCollection 2024 Dec 10. Mol Ther Nucleic Acids. 2024. PMID: 39494147 Free PMC article. Review. - OliTag-seq enhances in cellulo detection of CRISPR-Cas9 off-targets.
Yang ZX, Deng DH, Gao ZY, Zhang ZK, Fu YW, Wen W, Zhang F, Li X, Li HY, Zhang JP, Zhang XB. Yang ZX, et al. Commun Biol. 2024 Jun 6;7(1):696. doi: 10.1038/s42003-024-06360-w. Commun Biol. 2024. PMID: 38844522 Free PMC article. - Current Strategies for Increasing Knock-In Efficiency in CRISPR/Cas9-Based Approaches.
Leal AF, Herreno-Pachón AM, Benincore-Flórez E, Karunathilaka A, Tomatsu S. Leal AF, et al. Int J Mol Sci. 2024 Feb 20;25(5):2456. doi: 10.3390/ijms25052456. Int J Mol Sci. 2024. PMID: 38473704 Free PMC article. Review. - Precise genome-editing in human diseases: mechanisms, strategies and applications.
Zheng Y, Li Y, Zhou K, Li T, VanDusen NJ, Hua Y. Zheng Y, et al. Signal Transduct Target Ther. 2024 Feb 26;9(1):47. doi: 10.1038/s41392-024-01750-2. Signal Transduct Target Ther. 2024. PMID: 38409199 Free PMC article. Review. - Recent advances in CRISPR-Cas9-based genome insertion technologies.
Chen X, Du J, Yun S, Xue C, Yao Y, Rao S. Chen X, et al. Mol Ther Nucleic Acids. 2024 Feb 5;35(1):102138. doi: 10.1016/j.omtn.2024.102138. eCollection 2024 Mar 12. Mol Ther Nucleic Acids. 2024. PMID: 38379727 Free PMC article. Review.
References
- Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res. 2001;477:131–153. - PubMed
- Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous