Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection - PubMed (original) (raw)
Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection
Anna R Cliffe et al. J Virol. 2008 Dec.
Abstract
During lytic infection, the genome of herpes simplex virus 1 (HSV-1) is associated with limited levels of histones but does not form a regular repeating nucleosomal structure. However, the previous observation that chromatin remodeling factors are recruited into viral replication compartments indicates that chromatin remodeling plays a role in HSV-1 gene expression and DNA replication. In this study we demonstrate the presence of histone H3 on HSV-1 DNA early in infection at levels equivalent to those found on a cellular gene. The proportion of viral DNA associated with histone H3 decreases at later times postinfection, independently of either viral DNA replication or transcription. We demonstrate that an immediate-early protein, infected cell protein 0 (ICP0), is required for both a reduction in the proportion of HSV-1 DNA associating with histone H3 and an increase in histone acetylation. This study provides evidence that ICP0 directly alters the chromatin structure of the HSV-1 genome during lytic infection, and this system will serve as a useful model for the reduction of histone load in higher eukaryotes.
Figures
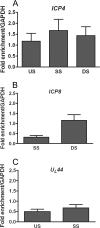
FIG. 1.
ChIP analysis of the fraction of HSV-1 DNA associated with histone H3. Total cell extracts were prepared at 3 hpi from HeLa cells infected with HSV-1 at an MOI of 0.1 PFU/cell. ChIP was carried out with an antibody specific to the C terminus of histone H3. The fraction of DNA immunoprecipitated compared to the input value was determined by real-time PCR, with the fraction immunoprecipitated by a nonspecific antibody subtracted, and is expressed as the fold enrichment over the fraction of GAPDH DNA immunoprecipitated. Primers were specific for regions of ICP4 (A), ICP8 (B), and UL44 (C) (see Table 1 for primer locations and sequences). Abbreviations: US, upstream of the transcriptional start site; SS, start site; DS, downstream of the transcriptional start site. The means and standard errors of the means of at least five independent experiments are shown.
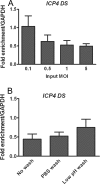
FIG. 2.
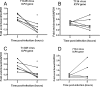
Effects of MOI and inactivation of nonadsorbed virus on the proportion of viral DNA associated with histone H3. ChIP was carried out at 3 hpi with an antibody for histone H3, and real-time time PCR was undertaken using primers for the region downstream of the ICP4 transcriptional start site and analyzed as for Fig. 1. (A) HeLa cells were infected at MOIs between 0.1 and 5 PFU/cell. (B) HeLa cells were infected at an MOI of 0.1 PFU/cell. After a 1-h adsorption period, the medium was replaced without washing the cells, or the cells were washed with PBS alone or washed with PBS followed by a low-pH wash buffer. The means and standard errors of the means of at least four independent experiments are shown.
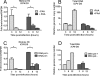
FIG. 3.
Kinetics of histone H3 association with the HSV genome and effects of viral DNA replication. HeLa cells were infected at an MOI of 0.1 PFU/cell in the presence or absence of the DNA polymerase inhibitor PAA (A and B) or with either a DNA Pol− virus or the rescued DNA Pol+ virus (C and D). Cell lysates were prepared at 3, 8, and 12 h postinfection. (A and C) ChIP was carried out using anti-histone H3 antibody. The fraction of ICP4 downstream (DS) (A) or ICP8 DS (C) DNA immunoprecipitated as determined by real-time PCR was normalized to the fraction of GAPDH DNA immunoprecipitated. The HSV genome copy number determined by real-time PCR was normalized to the GAPDH copy number and then normalized to the results for the untreated 3-h time point (B and D). The means and standard errors of the means of at least three independent experiments are shown. A significant decrease (*) between the mean fraction of viral DNA immunoprecipitated between 3 and 8 hpi was determined using Student's t test (P < 0.05).
FIG. 4.
Effects of DNA replication on the total amount of viral DNA associated with histone H3. HeLa cells were infected at an MOI of 0.1 PFU/cell in the presence or absence of the DNA polymerase inhibitor PAA. Cell lysates were prepared at 12 hpi. (A) ChIP assays were carried out using anti-histone H3 antibody. The copy number of ICP4 downstream DNA associated with histone H3 was determined by real-time PCR and normalized to the copy number of GAPDH associated with histone H3 and then normalized to the results for PAA-treated cells. (B) Total HSV genome copy number determined by real-time PCR using primers for the ICP4 gene was normalized to the GAPDH copy number and then normalized to results with the PAA-treated cells. The means and standard errors of the means are shown. Samples with mean values that vary significantly (P < 0.05, Student's t test) are indicated (*).
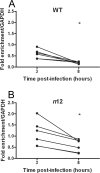
FIG. 5.
Kinetics of histone H3 association with the ICP8 gene following infection with an ICP4 nonsense mutant virus. HeLa cells were infected at an MOI of 0.1 PFU/cell with either WT (A) or ICP4− (B) virus. Cell lysates were prepared at 3 and 8 hpi. The fraction of ICP8 downstream DNA immunoprecipitated, as determined by real-time PCR, was normalized to the fraction of GAPDH DNA immunoprecipitated. Lines connect values for individual experiments. Samples with mean values that varied significantly (P < 0.05, paired Student's t test) are indicated (*).
FIG. 6.
Kinetics of histone H3 association with the ICP4 and ICP8 genes following infection with a ICP0− virus. HeLa cells were infected at an MOI of 0.1 PFU/cell with either an ICP0− virus (7134) (B and D) or rescued virus (7134R) (A and C). Cell lysates were prepared at 3 and 8 hpi. The fraction of ICP4 downstream (A and B) or ICP8 downstream (C and D) DNA immunoprecipitated determined by real-time PCR was normalized to the fraction of GAPDH DNA immunoprecipitated. Lines connect values for individual experiments. Samples with mean values that varied significantly (P < 0.05, paired Student's t test) are indicated (*).
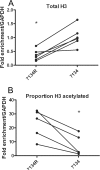
FIG. 7.
ChIP for total histone H3 and acetylated histone H3 on the ICP8 gene in the presence and absence of ICP0. HeLa cells were infected an MOI of 0.1 PFU/cell with either 7134 ICP0− virus or 7134R rescued virus (ICP0+). Cell lysates were prepared at 6 hpi, and ChIP was carried out using anti-histone H3 (A and B) and anti-acH3 (K9/18) (B) antibodies. The proportion of acetylated histone H3 associating with viral DNA was determined as a fraction of DNA associating with acH3 K9/14, normalized to the fraction associating with total histone H3. Lines connect values for individual experiments. Samples with mean values that varied significantly (P < 0.05, paired Student's t test) are indicated (*).
Similar articles
- Cellular Transcriptional Coactivator RanBP10 and Herpes Simplex Virus 1 ICP0 Interact and Synergistically Promote Viral Gene Expression and Replication.
Sato Y, Kato A, Maruzuru Y, Oyama M, Kozuka-Hata H, Arii J, Kawaguchi Y. Sato Y, et al. J Virol. 2016 Jan 6;90(6):3173-86. doi: 10.1128/JVI.03043-15. J Virol. 2016. PMID: 26739050 Free PMC article. - Herpes simplex virus VP16, but not ICP0, is required to reduce histone occupancy and enhance histone acetylation on viral genomes in U2OS osteosarcoma cells.
Hancock MH, Cliffe AR, Knipe DM, Smiley JR. Hancock MH, et al. J Virol. 2010 Feb;84(3):1366-75. doi: 10.1128/JVI.01727-09. Epub 2009 Nov 25. J Virol. 2010. PMID: 19939931 Free PMC article. - Herpes simplex virus type 1 ICP0 protein mediates activation of adeno-associated virus type 2 rep gene expression from a latent integrated form.
Geoffroy MC, Epstein AL, Toublanc E, Moullier P, Salvetti A. Geoffroy MC, et al. J Virol. 2004 Oct;78(20):10977-86. doi: 10.1128/JVI.78.20.10977-10986.2004. J Virol. 2004. PMID: 15452218 Free PMC article. - Chromatin dynamics during herpes simplex virus-1 lytic infection.
Placek BJ, Berger SL. Placek BJ, et al. Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):223-7. doi: 10.1016/j.bbagrm.2010.01.012. Epub 2010 Feb 6. Biochim Biophys Acta. 2010. PMID: 20139038 Review. - Role of chromatin during herpesvirus infections.
Kutluay SB, Triezenberg SJ. Kutluay SB, et al. Biochim Biophys Acta. 2009 Jun;1790(6):456-66. doi: 10.1016/j.bbagen.2009.03.019. Epub 2009 Mar 31. Biochim Biophys Acta. 2009. PMID: 19344747 Free PMC article. Review.
Cited by
- Entry of herpes simplex virus type 1 (HSV-1) into the distal axons of trigeminal neurons favors the onset of nonproductive, silent infection.
Hafezi W, Lorentzen EU, Eing BR, Müller M, King NJ, Klupp B, Mettenleiter TC, Kühn JE. Hafezi W, et al. PLoS Pathog. 2012;8(5):e1002679. doi: 10.1371/journal.ppat.1002679. Epub 2012 May 10. PLoS Pathog. 2012. PMID: 22589716 Free PMC article. - Daxx mediated histone H3.3 deposition on HSV-1 DNA restricts genome decompaction and the progression of immediate-early transcription.
Roberts APE, Orr A, Iliev V, Orr L, McFarlane S, Yang Z, Epifano I, Loney C, Rodriguez MC, Cliffe AR, Conn KL, Boutell C. Roberts APE, et al. bioRxiv [Preprint]. 2024 Aug 15:2024.08.15.608064. doi: 10.1101/2024.08.15.608064. bioRxiv. 2024. PMID: 39185184 Free PMC article. Preprint. - Use of biotinylated plasmid DNA as a surrogate for HSV DNA to identify proteins that repress or activate viral gene expression.
Mallon S, Wakim BT, Roizman B. Mallon S, et al. Proc Natl Acad Sci U S A. 2012 Dec 18;109(51):E3549-57. doi: 10.1073/pnas.1218783109. Epub 2012 Dec 5. Proc Natl Acad Sci U S A. 2012. PMID: 23223531 Free PMC article. - A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation.
Kennedy PG, Rovnak J, Badani H, Cohrs RJ. Kennedy PG, et al. J Gen Virol. 2015 Jul;96(Pt 7):1581-602. doi: 10.1099/vir.0.000128. Epub 2015 Mar 20. J Gen Virol. 2015. PMID: 25794504 Free PMC article. Review. - A Herpesviral Lytic Protein Regulates the Structure of Latent Viral Chromatin.
Raja P, Lee JS, Pan D, Pesola JM, Coen DM, Knipe DM. Raja P, et al. mBio. 2016 May 17;7(3):e00633-16. doi: 10.1128/mBio.00633-16. mBio. 2016. PMID: 27190217 Free PMC article.
References
- Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111381-392. - PubMed
- Bartova, E., J. Pachernik, A. Harnicarova, A. Kovarik, M. Kovarikova, J. Hofmanova, M. Skalnikova, M. Kozubek, and S. Kozubek. 2005. Nuclear levels and patterns of histone H3 modification and HP1 proteins after inhibition of histone deacetylases. J. Cell Sci. 1185035-5046. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources