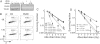Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability - PubMed (original) (raw)
Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability
Jennifer Blickwedehl et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Proteasome activator PA200 enhances proteasome-mediated cleavage after acidic residues in vitro; however, its role within cells is not known. Here, we show that, in response to ionizing radiation, PA200 forms hybrid proteasomes with 19S caps and 20S core proteasomes that accumulate on chromatin, leading to an increase in proteolytic activity. Unlike many other proteins that respond to DNA damage, the response of PA200 appears to be independent of Ataxia Telangiectasia Mutated and p53, but dependent on DNA-dependent protein kinase activity. Nonetheless, PA200 is critical because PA200-knockdown cells show genomic instability and reduced survival after exposure to ionizing radiation. This phenotype is reproduced by specific inhibition of postglutamyl activity of proteasomes, but combined treatment with PA200 siRNA and postglutamyl inhibitor does not show additive effects on survival. Together, these data suggest a unique role for PA200 in genomic stability that is likely mediated through its ability to enhance postglutamyl cleavage by proteasomes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
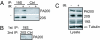
PA200-19S hybrid proteasomes are enhanced after IR exposure. Twenty-four hours after mock treatment (−) or 50-Gy irradiation (+) is shown. (A) Equivalent numbers of live HeLa cells were subjected to immunoprecipitation (IP) for a subunit (S5a) of the 19S cap (19S) or by using a control antibody (Ctrl). Bound proteins were immunoblotted with PA200-specific antiserum or MCP20 antibody (20S). (B) HeLa cells were immunoprecipitated (19S, 1st IP) as in A. Beads were eluted in 0.4 M NaCl and re-IP (2nd IP) for core proteasomes or by using a control antibody (Ctrl). Bound proteins were immunoblotted with PA200-specific antiserum. (C) Western blot analysis of lysates.
Fig. 2.
Increased hybrid proteasomes and proteasome activity on chromatin after IR exposure. (A) HeLa cells were mock treated (−) or exposed to 50-Gy IR (+), and chromatin-bound proteins were immunoblotted 24 h after IR exposure. (B) Normal BJ fibroblasts were irradiated with 5 Gy and assessed at various time points as in A. Each figure shown is representative of at least three independent experiments. (C and D) An aliquot of the chromatin-bound proteins from A were assessed for proteasome activity 24 h after IR exposure (*, P < 0.001). Specific fluorescence was calculated by subtraction of the fluorescence released in the presence of inhibitor from the absence of inhibitor. Error bars are SD of triplicates. (E and F) HeLa cells were transfected with control (Ctrl) or PA200-specific (PA200) siRNA. (E) Depletion of PA200 was confirmed by immunoblotting of cell lysates. (F) Cells were exposed to 50-Gy IR, and chromatin-bound proteins were assessed for postglutamyl proteolytic activity as described above.
Fig. 3.
PA200 accumulation on chromatin is independent of ATM but dependent on DNA-PK activity. Chromatin-bound proteins were immunoblotted for the indicated proteins. (A) HeLa cells were mock (−) or 50-Gy-irradiated (+) in the presence or absence of the ATM-specific inhibitor (ATMi) KU-55933 (10 μM) or the DNA-PK-specific inhibitor (PKi) NU7026 (10 μM). (B and C) The band intensities in A were quantified as fluorescence normalized to KU70 protein levels. (D) Normal BJ fibroblasts (WT) or ATM-null GM-5823 cells (null) were mock (−) or 5-Gy-irradiated (+) and analyzed after 24 h.
Fig. 4.
PA200 and postglutamyl peptide hydrolysis are essential for survival from IR-induced damage. (A–D) HeLa cells were transfected with control (Ctrl) siRNA, PA200-specific (PA200) siRNA, siRNA specific for the 3′-UTR of PA200 (UTR) with or without a PA200 cDNA expression plasmid. (A) Depletion of PA200 and restoration by PA200 cDNA was confirmed by immunoblotting of cell lysates. (B) Staining for phosphorylated histone H3 (_P_-H3) and DNA content (PI) of control siRNA and PA200 siRNA cells exposed to 50-Gy IR (+IR) or not (−IR). Percentage of gated cells within the indicated region (R3) is indicated for each siRNA and treatment. (C) Indicated cells were exposed to varying doses of IR and monitored for cell survival. Error bars indicate SD of three independent experiments. (*, P < 0.05, by χ2 analysis). (D) Indicated cells were treated with DMSO (Ctrl) or proteasome inhibitors (1 nM PS-341 or 1 μM YU-102) for 24 h and assessed as in C. Cells treated with YU-102 exhibit significantly reduced cell survival after IR exposure compared with control cells (**, P < 0.01, by χ2 analysis; *, P < 0.05). Error bars indicate SD from three independent experiments.
Similar articles
- Proteasomes and proteasome activator 200 kDa (PA200) accumulate on chromatin in response to ionizing radiation.
Blickwedehl J, McEvoy S, Wong I, Kousis P, Clements J, Elliott R, Cresswell P, Liang P, Bangia N. Blickwedehl J, et al. Radiat Res. 2007 Jun;167(6):663-74. doi: 10.1667/RR0690.1. Radiat Res. 2007. PMID: 17523843 - Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability.
Latonen L, Moore HM, Bai B, Jäämaa S, Laiho M. Latonen L, et al. Oncogene. 2011 Feb 17;30(7):790-805. doi: 10.1038/onc.2010.469. Epub 2010 Oct 18. Oncogene. 2011. PMID: 20956947 - Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures.
Guan H, Wang Y, Yu T, Huang Y, Li M, Saeed AFUH, Perčulija V, Li D, Xiao J, Wang D, Zhu P, Ouyang S. Guan H, et al. PLoS Biol. 2020 Mar 5;18(3):e3000654. doi: 10.1371/journal.pbio.3000654. eCollection 2020 Mar. PLoS Biol. 2020. PMID: 32134919 Free PMC article. - The Proteasome Activator PA200/PSME4: An Emerging New Player in Health and Disease.
Yazgili AS, Ebstein F, Meiners S. Yazgili AS, et al. Biomolecules. 2022 Aug 20;12(8):1150. doi: 10.3390/biom12081150. Biomolecules. 2022. PMID: 36009043 Free PMC article. Review. - Proteasome activator 200: the heat is on..
Savulescu AF, Glickman MH. Savulescu AF, et al. Mol Cell Proteomics. 2011 May;10(5):R110.006890. doi: 10.1074/mcp.R110.006890. Epub 2011 Mar 9. Mol Cell Proteomics. 2011. PMID: 21389348 Free PMC article. Review.
Cited by
- Histone degradation by the proteasome regulates chromatin and cellular plasticity.
Shmueli MD, Sheban D, Eisenberg-Lerner A, Merbl Y. Shmueli MD, et al. FEBS J. 2022 Jun;289(12):3304-3316. doi: 10.1111/febs.15903. Epub 2021 May 13. FEBS J. 2022. PMID: 33914417 Free PMC article. Review. - Ubiquitin/proteasome pathway impairment in neurodegeneration: therapeutic implications.
Huang Q, Figueiredo-Pereira ME. Huang Q, et al. Apoptosis. 2010 Nov;15(11):1292-311. doi: 10.1007/s10495-010-0466-z. Apoptosis. 2010. PMID: 20131003 Free PMC article. Review. - Jab1 promotes gastric cancer tumorigenesis via non-ubiquitin proteasomal degradation of p14ARF.
Wang L, Du WQ, Xie M, Liu MR, Huo FC, Yang J, Pei DS. Wang L, et al. Gastric Cancer. 2020 Nov;23(6):1003-1017. doi: 10.1007/s10120-020-01087-z. Epub 2020 May 26. Gastric Cancer. 2020. PMID: 32458234 - Developing a Bimolecular Affinity Purification Strategy to Isolate 26S Proteasome Holocomplexes for Complex-Centric Proteomic Analysis.
Yu C, Wang X, Li W, Liu Y, Huang L. Yu C, et al. Anal Chem. 2021 Oct 5;93(39):13407-13413. doi: 10.1021/acs.analchem.1c03551. Epub 2021 Sep 22. Anal Chem. 2021. PMID: 34550675 Free PMC article. - The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response.
Butler LR, Densham RM, Jia J, Garvin AJ, Stone HR, Shah V, Weekes D, Festy F, Beesley J, Morris JR. Butler LR, et al. EMBO J. 2012 Oct 3;31(19):3918-34. doi: 10.1038/emboj.2012.232. Epub 2012 Aug 21. EMBO J. 2012. PMID: 22909820 Free PMC article.
References
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol Rev. 2002;82:373–428. - PubMed
- Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: Biological regulation via destruction. Bioessays. 2000;22:442–451. - PubMed
- Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: Cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. - PubMed
- Khanna KK, Jackson SP. DNA double-strand breaks: Signaling, repair, and the cancer connection. Nat Genet. 2001;27:247–254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016056/CA/NCI NIH HHS/United States
- CA10445/CA/NCI NIH HHS/United States
- CA123232/CA/NCI NIH HHS/United States
- CA085183/CA/NCI NIH HHS/United States
- R01 CA123232/CA/NCI NIH HHS/United States
- T32 CA085183/CA/NCI NIH HHS/United States
- CA016056/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous