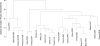Analysis of gene regulatory networks in the mammalian circadian rhythm - PubMed (original) (raw)
Comparative Study
Analysis of gene regulatory networks in the mammalian circadian rhythm
Jun Yan et al. PLoS Comput Biol. 2008 Oct.
Abstract
Circadian rhythm is fundamental in regulating a wide range of cellular, metabolic, physiological, and behavioral activities in mammals. Although a small number of key circadian genes have been identified through extensive molecular and genetic studies in the past, the existence of other key circadian genes and how they drive the genomewide circadian oscillation of gene expression in different tissues still remains unknown. Here we try to address these questions by integrating all available circadian microarray data in mammals. We identified 41 common circadian genes that showed circadian oscillation in a wide range of mouse tissues with a remarkable consistency of circadian phases across tissues. Comparisons across mouse, rat, rhesus macaque, and human showed that the circadian phases of known key circadian genes were delayed for 4-5 hours in rat compared to mouse and 8-12 hours in macaque and human compared to mouse. A systematic gene regulatory network for the mouse circadian rhythm was constructed after incorporating promoter analysis and transcription factor knockout or mutant microarray data. We observed the significant association of cis-regulatory elements: EBOX, DBOX, RRE, and HSE with the different phases of circadian oscillating genes. The analysis of the network structure revealed the paths through which light, food, and heat can entrain the circadian clock and identified that NR3C1 and FKBP/HSP90 complexes are central to the control of circadian genes through diverse environmental signals. Our study improves our understanding of the structure, design principle, and evolution of gene regulatory networks involved in the mammalian circadian rhythm.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
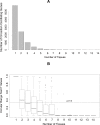
Figure 1. Tissue distribution of circadian oscillating genes.
(A) Distribution of the number of circadian oscillating genes identified in different numbers of mouse tissues. (B) Distribution of _p_-values in circular range tests for circadian phases of circadian oscillating genes identified in different numbers of mouse tissues.
Figure 2. Hierarchical clustering of 21 circadian microarray datasets based on global circadian phase dissimilarities.
Datasets are denoted by first author names and tissue types.
Figure 3. Comparison of circadian phases between SCN and liver.
_p_-values from the circular ANOVA test are indicated in the parenthesis. The solid line represents y = x. The dashed lines represent y = _x_±6 respectively.
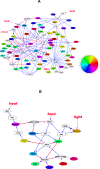
Figure 4. Circadian gene regulatory network in mouse.
(A) Gene regulatory network consisting of the circadian oscillating genes identified in at least 7 mouse tissues. (B) The subset of network highlighting NR3C1 and FKBP/HSP90's role of integrating the regulatory inputs from diverse environmental signals into circadian genes. Blue arrows represent the gene regulatory interactions obtained in this study. Red arrows represent the known gene regulatory or protein interactions extracted from the literature. P stands for phosphorylation. White boxes represent _cis-_regulatory elements. Colored circles represent the genes with circadian phase information, where circadian phases are represented by the different colors in the color wheel. White circles represent protein complexes or genes without circadian phase information.
Similar articles
- A network biology study on circadian rhythm by integrating various omics data.
Wang Y, Zhang XS, Chen L. Wang Y, et al. OMICS. 2009 Aug;13(4):313-24. doi: 10.1089/omi.2009.0040. OMICS. 2009. PMID: 19645592 - Transcriptional oscillation of canonical clock genes in mouse peripheral tissues.
Yamamoto T, Nakahata Y, Soma H, Akashi M, Mamine T, Takumi T. Yamamoto T, et al. BMC Mol Biol. 2004 Oct 9;5:18. doi: 10.1186/1471-2199-5-18. BMC Mol Biol. 2004. PMID: 15473909 Free PMC article. - Promoter analysis of Mammalian clock controlled genes.
Bozek K, Kiełbasa SM, Kramer A, Herzel H. Bozek K, et al. Genome Inform. 2007;18:65-74. Genome Inform. 2007. PMID: 18546475 - Chronobiology of micturition: putative role of the circadian clock.
Negoro H, Kanematsu A, Yoshimura K, Ogawa O. Negoro H, et al. J Urol. 2013 Sep;190(3):843-9. doi: 10.1016/j.juro.2013.02.024. Epub 2013 Feb 18. J Urol. 2013. PMID: 23429068 Review. - Molecular genetics of circadian rhythms in mammals.
King DP, Takahashi JS. King DP, et al. Annu Rev Neurosci. 2000;23:713-42. doi: 10.1146/annurev.neuro.23.1.713. Annu Rev Neurosci. 2000. PMID: 10845079 Review.
Cited by
- A survey of genomic studies supports association of circadian clock genes with bipolar disorder spectrum illnesses and lithium response.
McCarthy MJ, Nievergelt CM, Kelsoe JR, Welsh DK. McCarthy MJ, et al. PLoS One. 2012;7(2):e32091. doi: 10.1371/journal.pone.0032091. Epub 2012 Feb 22. PLoS One. 2012. PMID: 22384149 Free PMC article. - Circadian variations of clock gene Per2 and cell cycle genes in different stages of carcinogenesis in golden hamster buccal mucosa.
Tan XM, Ye H, Yang K, Chen D, Wang QQ, Tang H, Zhao NB. Tan XM, et al. Sci Rep. 2015 May 7;5:9997. doi: 10.1038/srep09997. Sci Rep. 2015. PMID: 25950458 Free PMC article. - The pervasiveness and plasticity of circadian oscillations: the coupled circadian-oscillators framework.
Patel VR, Ceglia N, Zeller M, Eckel-Mahan K, Sassone-Corsi P, Baldi P. Patel VR, et al. Bioinformatics. 2015 Oct 1;31(19):3181-8. doi: 10.1093/bioinformatics/btv353. Epub 2015 Jun 6. Bioinformatics. 2015. PMID: 26049162 Free PMC article. - Circadian patterns of gene expression in the human brain and disruption in major depressive disorder.
Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, Evans SJ, Choudary PV, Cartagena P, Barchas JD, Schatzberg AF, Jones EG, Myers RM, Watson SJ Jr, Akil H, Bunney WE. Li JZ, et al. Proc Natl Acad Sci U S A. 2013 Jun 11;110(24):9950-5. doi: 10.1073/pnas.1305814110. Epub 2013 May 13. Proc Natl Acad Sci U S A. 2013. PMID: 23671070 Free PMC article. - The endogenous molecular clock orchestrates the temporal separation of substrate metabolism in skeletal muscle.
Hodge BA, Wen Y, Riley LA, Zhang X, England JH, Harfmann BD, Schroder EA, Esser KA. Hodge BA, et al. Skelet Muscle. 2015 May 16;5:17. doi: 10.1186/s13395-015-0039-5. eCollection 2015. Skelet Muscle. 2015. PMID: 26000164 Free PMC article.
References
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. - PubMed
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. - PubMed
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. - PubMed
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous