Chromatin insulators: regulatory mechanisms and epigenetic inheritance - PubMed (original) (raw)
Review
Chromatin insulators: regulatory mechanisms and epigenetic inheritance
Ashley M Bushey et al. Mol Cell. 2008.
Abstract
Enhancer-blocking insulators are DNA elements that disrupt the communication between a regulatory sequence, such as an enhancer or a silencer, and a promoter. Insulators participate in both transcriptional regulation and global nuclear organization, two features of chromatin that are thought to be maintained from one generation to the next through epigenetic mechanisms. Furthermore, there are many regulatory mechanisms in place that enhance or hinder insulator activity. These modes of regulation could be used to establish cell-type-specific insulator activity that is epigenetically inherited along a cell and/or organismal lineage. This review will discuss the evidence for epigenetic inheritance and regulation of insulator function.
Figures
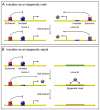
Figure 1
The role of insulators in epigenetic memory. (A) Insulator protein binding could be an epigenetic mark that is maintained through cell division. Locus A and Locus B represent two different genomic loci that contain insulator elements. Locus A is bound by an insulator protein that prevents transcription, whereas locus B is unbound and therefore able to produce a gene product (pentagon). If insulator protein binding acts as an epigenetic mark, binding or lack of binding is remembered after cell division. (B) An insulator could participate in an epigenetic signal through regulating a gene that establishes an epigenetic mark. In this situation, loss of insulator protein binding leads to production of a gene product (pentagon) that establishes an epigenetic mark, here in Locus B. The epigenetic mark is maintained through cell division regardless of the state of the insulator. Therefore, the insulator participates in the establishment of the epigenetic mark, but is not the mark itself. Squares – enhancer binding proteins; Circles – insulator proteins; Pentagons – gene products whose expression is regulated by insulators; Ovals – epigenetic marks.
Figure 2
Regulation of insulator activity. (A) Insulator function could be regulated on the level of DNA-binding activity. Regulation could be positive or negative and could occur by various molecular mechanisms, including DNA modification, protein modification, protein-protein interaction, and effects of nearby chromatin. (B) Insulator function could be regulated on the level of protein-protein interaction. Again, regulation could be positive or negative. Molecular mechanisms could include protein modification, protein-protein interaction, effects of nearby chromatin, and interaction with small RNAs.
Similar articles
- Chromatin insulators.
Valenzuela L, Kamakaka RT. Valenzuela L, et al. Annu Rev Genet. 2006;40:107-38. doi: 10.1146/annurev.genet.39.073003.113546. Annu Rev Genet. 2006. PMID: 16953792 Review. - A functional insulator screen identifies NURF and dREAM components to be required for enhancer-blocking.
Bohla D, Herold M, Panzer I, Buxa MK, Ali T, Demmers J, Krüger M, Scharfe M, Jarek M, Bartkuhn M, Renkawitz R. Bohla D, et al. PLoS One. 2014 Sep 23;9(9):e107765. doi: 10.1371/journal.pone.0107765. eCollection 2014. PLoS One. 2014. PMID: 25247414 Free PMC article. - Impact of Interactions between Su(Hw)-Dependent Insulators on the Transvection Effect in Drosophila melanogaster.
Melnikova LS, Molodina VV, Georgiev PG, Golovnin AK. Melnikova LS, et al. Dokl Biochem Biophys. 2024 Aug;517(1):127-133. doi: 10.1134/S1607672924700820. Epub 2024 May 14. Dokl Biochem Biophys. 2024. PMID: 38744735 - Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein.
Parnell TJ, Kuhn EJ, Gilmore BL, Helou C, Wold MS, Geyer PK. Parnell TJ, et al. Mol Cell Biol. 2006 Aug;26(16):5983-93. doi: 10.1128/MCB.00698-06. Mol Cell Biol. 2006. PMID: 16880510 Free PMC article. - Functional sub-division of the Drosophila genome via chromatin looping: the emerging importance of CP190.
Ahanger SH, Shouche YS, Mishra RK. Ahanger SH, et al. Nucleus. 2013 Mar-Apr;4(2):115-22. doi: 10.4161/nucl.23389. Epub 2013 Jan 18. Nucleus. 2013. PMID: 23333867 Free PMC article. Review.
Cited by
- Repetitive elements and enforced transcriptional repression co-operate to enhance DNA methylation spreading into a promoter CpG-island.
Zhang Y, Shu J, Si J, Shen L, Estecio MR, Issa JP. Zhang Y, et al. Nucleic Acids Res. 2012 Aug;40(15):7257-68. doi: 10.1093/nar/gks429. Epub 2012 May 17. Nucleic Acids Res. 2012. PMID: 22600741 Free PMC article. - Loss of the insulator protein CTCF during nematode evolution.
Heger P, Marin B, Schierenberg E. Heger P, et al. BMC Mol Biol. 2009 Aug 27;10:84. doi: 10.1186/1471-2199-10-84. BMC Mol Biol. 2009. PMID: 19712444 Free PMC article. - Insulators as mediators of intra- and inter-chromosomal interactions: a common evolutionary theme.
Ong CT, Corces VG. Ong CT, et al. J Biol. 2009 Aug 27;8(8):73. doi: 10.1186/jbiol165. J Biol. 2009. PMID: 19725934 Free PMC article. Review. - Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease.
Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Larivière M, Moussette S, Grundberg E, Kwan T, Ouimet M, Ge B, Hoberman R, Swiatek M, Dias J, Lam KC, Koka V, Harmsen E, Soto-Quiros M, Avila L, Celedón JC, Weiss ST, Dewar K, Sinnett D, Laprise C, Raby BA, Pastinen T, Naumova AK. Verlaan DJ, et al. Am J Hum Genet. 2009 Sep;85(3):377-93. doi: 10.1016/j.ajhg.2009.08.007. Am J Hum Genet. 2009. PMID: 19732864 Free PMC article. - Stalled Hox promoters as chromosomal boundaries.
Chopra VS, Cande J, Hong JW, Levine M. Chopra VS, et al. Genes Dev. 2009 Jul 1;23(13):1505-9. doi: 10.1101/gad.1807309. Epub 2009 Jun 10. Genes Dev. 2009. PMID: 19515973 Free PMC article.
References
- Awad TA, Bigler J, Ulmer JE, Hu YJ, Moore JM, Lutz M, Neiman PE, Collins SJ, Renkawitz R, Lobanenkov VV, Filippova GN. Negative transcriptional regulation mediated by thyroid hormone response element 144 requires binding of the multivalent factor CTCF to a novel target DNA sequence. The Journal of biological chemistry. 1999;274:27092–27098. - PubMed
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases