Fast phylogenetic DNA barcoding - PubMed (original) (raw)
Fast phylogenetic DNA barcoding
Kasper Munch et al. Philos Trans R Soc Lond B Biol Sci. 2008.
Abstract
We present a heuristic approach to the DNA assignment problem based on phylogenetic inferences using constrained neighbour joining and non-parametric bootstrapping. We show that this method performs as well as the more computationally intensive full Bayesian approach in an analysis of 500 insect DNA sequences obtained from GenBank. We also analyse a previously published dataset of environmental DNA sequences from soil from New Zealand and Siberia, and use these data to illustrate the fact that statistical approaches to the DNA assignment problem allow for more appropriate criteria for determining the taxonomic level at which a particular DNA sequence can be assigned.
Figures
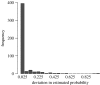
Figure 1
Histogram showing the difference in probabilities of assignment to the correct species estimated using the neighbour joining and MCMC.
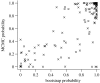
Figure 2
Estimated probabilities of assignment to the correct species using neighbour joining are plotted against the estimate obtained using MCMC.
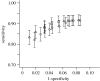
Figure 3
ROC curves summarizing the trade-off between sensitivity and specificity in the range of most to least stringent assignment criteria used. Sensitivity is the fraction of all sequences that are correctly assigned and specificity is the fraction of assignments that are correct. Vertical bars represent confidence intervals of the sensitivity statistic. Triangles, NJ; circles, MCMC.
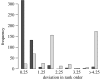
Figure 4
Histogram illustrating the agreement in terms of rank order obtained by sorting the set of homologues by the assignment probability associated obtained neighbour joining with bootstrapping and maximum likelihood and MCMC. The histograms show the average difference in rank order for neighbour joining and BLAST from the one obtained using MCMC.
Similar articles
- Statistical assignment of DNA sequences using Bayesian phylogenetics.
Munch K, Boomsma W, Huelsenbeck JP, Willerslev E, Nielsen R. Munch K, et al. Syst Biol. 2008 Oct;57(5):750-7. doi: 10.1080/10635150802422316. Syst Biol. 2008. PMID: 18853361 - A fuzzy-set-theory-based approach to analyse species membership in DNA barcoding.
Zhang AB, Muster C, Liang HB, Zhu CD, Crozier R, Wan P, Feng J, Ward RD. Zhang AB, et al. Mol Ecol. 2012 Apr;21(8):1848-63. doi: 10.1111/j.1365-294X.2011.05235.x. Epub 2011 Aug 29. Mol Ecol. 2012. PMID: 21883585 - Hyperbolic SOM-based clustering of DNA fragment features for taxonomic visualization and classification.
Martin C, Diaz NN, Ontrup J, Nattkemper TW. Martin C, et al. Bioinformatics. 2008 Jul 15;24(14):1568-74. doi: 10.1093/bioinformatics/btn257. Epub 2008 Jun 5. Bioinformatics. 2008. PMID: 18535082 - DNA barcoding in plants: evolution and applications of in silico approaches and resources.
Bhargava M, Sharma A. Bhargava M, et al. Mol Phylogenet Evol. 2013 Jun;67(3):631-41. doi: 10.1016/j.ympev.2013.03.002. Epub 2013 Mar 13. Mol Phylogenet Evol. 2013. PMID: 23500333 Review. - Land plants and DNA barcodes: short-term and long-term goals.
Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP, Haider N, Savolainen V. Chase MW, et al. Philos Trans R Soc Lond B Biol Sci. 2005 Oct 29;360(1462):1889-95. doi: 10.1098/rstb.2005.1720. Philos Trans R Soc Lond B Biol Sci. 2005. PMID: 16214746 Free PMC article. Review.
Cited by
- The microbiome and metatranscriptome of a panel from the Sarracenia mapping population reveal complex assembly and function involving host influence.
Cai J, Mohsin I, Rogers W, Zhang M, Jiang L, Malmberg R, Alabady M. Cai J, et al. Front Plant Sci. 2024 Oct 15;15:1445713. doi: 10.3389/fpls.2024.1445713. eCollection 2024. Front Plant Sci. 2024. PMID: 39474216 Free PMC article. - DEPP: Deep Learning Enables Extending Species Trees using Single Genes.
Jiang Y, Balaban M, Zhu Q, Mirarab S. Jiang Y, et al. Syst Biol. 2023 May 19;72(1):17-34. doi: 10.1093/sysbio/syac031. Syst Biol. 2023. PMID: 35485976 Free PMC article. - Biodiversity inventory of the grey mullets (Actinopterygii: Mugilidae) of the Indo-Australian Archipelago through the iterative use of DNA-based species delimitation and specimen assignment methods.
Delrieu-Trottin E, Durand JD, Limmon G, Sukmono T, Kadarusman, Sugeha HY, Chen WJ, Busson F, Borsa P, Dahruddin H, Sauri S, Fitriana Y, Zein MSA, Hocdé R, Pouyaud L, Keith P, Wowor D, Steinke D, Hanner R, Hubert N. Delrieu-Trottin E, et al. Evol Appl. 2020 Feb 11;13(6):1451-1467. doi: 10.1111/eva.12926. eCollection 2020 Jul. Evol Appl. 2020. PMID: 32684969 Free PMC article. - Integrative Profiling of Bee Communities from Habitats of Tropical Southern Yunnan (China).
Liu XW, Chesters D, Dai QY, Niu ZQ, Beckschäfer P, Martin K, Zhu CD. Liu XW, et al. Sci Rep. 2017 Jul 13;7(1):5336. doi: 10.1038/s41598-017-05262-8. Sci Rep. 2017. PMID: 28706192 Free PMC article. - Unidentifiable by morphology: DNA barcoding of plant material in local markets in Iran.
Ghorbani A, Saeedi Y, de Boer HJ. Ghorbani A, et al. PLoS One. 2017 Apr 18;12(4):e0175722. doi: 10.1371/journal.pone.0175722. eCollection 2017. PLoS One. 2017. PMID: 28419161 Free PMC article.
References
- Abdo Z, Golding G.B. A step toward barcoding life: a model-based, decision-theoretic method to assign genes to preexisting species groups. Syst. Biol. 2007;56:44–56. doi:10.1080/10635150601167005 - DOI - PubMed
- Alfaro M.E, Zoller S, Lutzoni F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Biol. Evol. 2003;20:255–266. doi:10.1093/molbev/msg028 - DOI - PubMed
- Dawnay N, Ogden R, McEwing R, Carvalho G.R, Thorpe R.S. Validation of the barcoding gene COI for use in forensic genetic species identification. Forensic Sci. Int. 2007;173:1–6. doi:10.1016/j.forsciint.2006.09.013 - DOI - PubMed
- Desper R, Gascuel O. Theoretical foundation of the balanced minimum evolution method of phylogenetic inference and its relationship to weighted least-squares tree fitting. Mol. Biol. Evol. 2004;21:587–598. doi:10.1093/molbev/msh049 - DOI - PubMed
- Douady C.J, Delsuc F, Boucher Y, Ford Doolittle W, Douzery E.J.P. Comparison of Bayesian and maximum likelihood bootstrap measures of phylogenetic reliability. Mol. Biol. Evol. 2003;20:248–252. doi:10.1093/molbev/msg042 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources