Biological principles of microRNA-mediated regulation: shared themes amid diversity - PubMed (original) (raw)
Review
Biological principles of microRNA-mediated regulation: shared themes amid diversity
Alex S Flynt et al. Nat Rev Genet. 2008 Nov.
Abstract
Regulation of gene activity by microRNAs is critical to myriad aspects of eukaryotic development and physiology. Amidst an extensive regulatory web that is predicted to involve thousands of transcripts, emergent themes are now beginning to illustrate how microRNAs have been incorporated into diverse settings. These include potent inhibition of individual key targets, fine-tuning of target activity, the coordinated regulation of target batteries, and the reversibility of some aspects of microRNA-mediated repression. Such themes may reflect some of the inherent advantages of exploiting microRNA control in biological circuits, and provide insight into the consequences of microRNA dysfunction in disease.
Figures
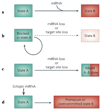
Figure 1. Classifying microRNA (miRNA) activities and functions
a The activity of a given target can be evaluated in the presence of a cognate miRNA. ‘Switch’ targets are essentially inactive following miRNA-mediated repression, whereas ‘tuning’ targets produce functional protein in the domain of miRNA activity. Both of these terms apply to evolutionarily conserved target relationships. Non-conserved but functional sites may mediate repression that is incidental or otherwise compensated for by other regulatory mechanisms; these are termed ‘neutral’ targets. Non-conserved sites may occasionally mediate beneficial, species-specific regulation. Not shown are ‘anti-targets’, transcripts for which miRNA-mediated repression is deterimental, such that the acquistion of target sites is avoided. b Another means of classification is to evaluate whether loss of function of a specific miRNA is associated with a phenotype, or whether the removal of target sites from a specific target causes a phenotype. For many conserved miRNAs and their conserved targets, the repression is likely to be beneficial enough to be selected during evolution but, overall, to be auxiliary to other means of gene regulation. Only a subset of miRNAs, with a small number of targets, mediate regulation with genetically defined consequences; we call these ‘miRNA genetic switches’, c One may consider the total number of targets whose repression by a miRNA in a given setting can be detected by quantitative means. Alternatively, a more stringent classification would be to consider only those targets whose repression by a miRNA is of detectable phenotypic consequence.
Figure 2. Consequences of microRNA (miRNA) loss- or gain-of-function for cell state
The activation of a miRNA can direct or aid a transition from state A to state B (a). Loss of a miRNA, or of its binding sites in a target, can lead to the cell becoming stuck in state A (b) or can result in a mixed state with characteristics of both state A and state B (c). An ectopic miRNA can prematurely induce state B or can cause cells to overcommit to this state (d).
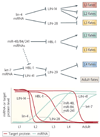
Figure 3. Multiple microRNA (miRNA) genetic switches in the nematode heterochronic pathway
The lin-4 miRNA promotes the transition between the first and second larval stages (L1 and L2, respectively) by repressing the transcription factor LIN-14 and the RNA-binding protein LIN-28. Similarly, three related miRNAs with homology to let-7 (miR-48, miR-84 and miR-241) act in concert to repress the transcription factor HBL-1, promoting the transition from L2 to L3. Finally, let-7 represses the TRIM protein LIN-41, and possibly HBL-1, to promote the transition from L4 to the adult.
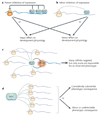
Figure 4. Configurations of microRNA (miRNA)–target networks
a Potent inhibition by a miRNA can have a substantial phenotypic effect in some cases, but can lead to only minor phenotypic consequences in other cases. b Relatively weak repression by a miRNA might often have minor consequences, but could have substantial effects if the biological system is sensitive to minor fluctuations in target activity. c Although most animal miRNAs are likely to influence a battery of targets in a given cell, the phenotype caused by loss of a miRNA might demonstrably be due to one or a few targets. d In some cases a miRNA might exert relatively equivalent effects across a large number of targets, the collective deregulation of which might lead to a detectable phenotype. However, the loss of miRNA-mediated repression of an entire target battery can also be fully compensated for by other regulatory mechanisms. RISC, RNA-induced silencing complex.
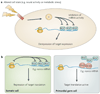
Figure 5. Antagonism of microRNA (miRNA)-mediated repression
a Signal-dependent alleviation of miRNA-mediated repression — for example, at synapses following neural activity, or in response to intracellular signals — might also trigger target re-activation, b Cell-type-specific RNA-binding proteins can antagonize miRNA activity For example, in the germ line, dead end 1 (DND1) can prevent the interaction between a miRNA and targets such as the nanos mRNA. RISC, RNA-induced silencing complex.
Similar articles
- MicroRNA networks and developmental plasticity in plants.
Rubio-Somoza I, Weigel D. Rubio-Somoza I, et al. Trends Plant Sci. 2011 May;16(5):258-64. doi: 10.1016/j.tplants.2011.03.001. Epub 2011 Apr 4. Trends Plant Sci. 2011. PMID: 21466971 Review. - Understanding microRNA-mediated gene regulatory networks through mathematical modelling.
Lai X, Wolkenhauer O, Vera J. Lai X, et al. Nucleic Acids Res. 2016 Jul 27;44(13):6019-35. doi: 10.1093/nar/gkw550. Epub 2016 Jun 17. Nucleic Acids Res. 2016. PMID: 27317695 Free PMC article. - MicroRNAs: key participants in gene regulatory networks.
Ke XS, Liu CM, Liu DP, Liang CC. Ke XS, et al. Curr Opin Chem Biol. 2003 Aug;7(4):516-23. doi: 10.1016/s1367-5931(03)00075-9. Curr Opin Chem Biol. 2003. PMID: 12941428 Review. - A conserved microRNA signal specifies leaf polarity.
Timmermans MC, Juarez MT, Phelps-Durr TL. Timmermans MC, et al. Cold Spring Harb Symp Quant Biol. 2004;69:409-17. doi: 10.1101/sqb.2004.69.409. Cold Spring Harb Symp Quant Biol. 2004. PMID: 16117675 Review. No abstract available. - Weak Regulation of Many Targets Is Cumulatively Powerful-An Evolutionary Perspective on microRNA Functionality.
Zhao Y, Shen X, Tang T, Wu CI. Zhao Y, et al. Mol Biol Evol. 2017 Dec 1;34(12):3041-3046. doi: 10.1093/molbev/msx260. Mol Biol Evol. 2017. PMID: 29029299 Review.
Cited by
- Bone sialoprotein facilitates anoikis resistance in lung cancer by inhibiting miR-150-5p expression.
Thuong LHH, Huang CL, Fong YC, Liu CL, Guo JH, Wu CY, Liu PI, Tang CH. Thuong LHH, et al. J Cell Mol Med. 2024 Oct;28(20):e70155. doi: 10.1111/jcmm.70155. J Cell Mol Med. 2024. PMID: 39466654 Free PMC article. - Mesenchymal stromal cell transplantation ameliorates fibrosis and microRNA dysregulation in skeletal muscle ischemia.
Sanz-Nogués C, Keane AJ, Creane M, Hynes SO, Chen X, Lyons CJ, Horan E, Elliman SJ, Goljanek-Whysall K, O'Brien T. Sanz-Nogués C, et al. Stem Cells. 2024 Nov 5;42(11):976-991. doi: 10.1093/stmcls/sxae058. Stem Cells. 2024. PMID: 39283740 Free PMC article. - Global-local aware Heterogeneous Graph Contrastive Learning for multifaceted association prediction in miRNA-gene-disease networks.
Si Y, Huang Z, Fang Z, Yuan Z, Huang Z, Li Y, Wei Y, Wu F, Yao YF. Si Y, et al. Brief Bioinform. 2024 Jul 25;25(5):bbae443. doi: 10.1093/bib/bbae443. Brief Bioinform. 2024. PMID: 39256197 Free PMC article. - In search of epigenetic hallmarks of different tissues: an integrative omics study of horse liver, lung, and heart.
Semik-Gurgul E, Pawlina-Tyszko K, Gurgul A, Szmatoła T, Rybińska J, Ząbek T. Semik-Gurgul E, et al. Mamm Genome. 2024 Dec;35(4):600-620. doi: 10.1007/s00335-024-10057-0. Epub 2024 Aug 14. Mamm Genome. 2024. PMID: 39143382 Free PMC article. - Coding and Non-Coding Transcriptomic Landscape of Aortic Complications in Marfan Syndrome.
Udugampolage NS, Frolova S, Taurino J, Pini A, Martelli F, Voellenkle C. Udugampolage NS, et al. Int J Mol Sci. 2024 Jul 5;25(13):7367. doi: 10.3390/ijms25137367. Int J Mol Sci. 2024. PMID: 39000474 Free PMC article. Review.
References
- Lau N, Lim L, Weinstein E, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. - PubMed
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. - PubMed
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. - PubMed
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. - PubMed
- Wightman B, Burglin TR, Gatto J, Arasu P, Ruvkun G. Negative regulatory sequences in the lin-14 3′-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 1991;5:1813–1824. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources