A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints - PubMed (original) (raw)
A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints
Xiajun Li et al. Dev Cell. 2008 Oct.
Abstract
The mechanisms responsible for maintaining genomic methylation imprints in mouse embryos are not understood. We generated a knockout mouse in the Zfp57 locus encoding a KRAB zinc finger protein. Loss of just the zygotic function of Zfp57 causes partial neonatal lethality, whereas eliminating both the maternal and zygotic functions of Zfp57 results in a highly penetrant embryonic lethality. In oocytes, absence of Zfp57 results in failure to establish maternal methylation imprints at the Snrpn imprinted region. Intriguingly, methylation imprints are reacquired specifically at the maternally derived Snrpn imprinted region when the zygotic Zfp57 is present in embryos. This suggests that there may be DNA methylation-independent memory for genomic imprints. Zfp57 is also required for the postfertilization maintenance of maternal and paternal methylation imprints at multiple imprinted domains. The effects on genomic imprinting are consistent with the maternal-zygotic lethality of Zfp57 mutants.
Figures
Figure 1. ZFP57 is a putative KRAB zinc finger protein
(A) Schematic diagram of the ZFP57 protein. (B) Sequence alignment between the mouse ZFP57 and the human ZFP57. The identical amino acids are boxed in black and the similar residues are shaded. (C) Co-IP assays were carried out for KAP-1 and myc epitope-tagged ZFP57. Three left lanes are the western blot of the total cell lysate (Input) from the KAP-1-transfected (lane 1), the KAP-1 and myc epitope-tagged ZFP57 co-transfected (lane 2) and the untransfected cos cells (lane 3). Lane 4, 5, 6 are the western blot of the immunoprecipitate (IP) derived from these three samples when the antibodies against myc epitope were used to pull down ZFP57-associated proteins. The rabbit polyclonal antibodies used for the western blot are Ab1 (anti-KAP1 RBCC) and Ab2 (anti-KAP1 CT) (Schultz et al., 2001). (D) Adult mouse organ Northern blot of Zfp57. A 1.2kb cDNA fragment encompassing the entire open-reading-frame of Zfp57 was used to probe a Northern blot. Equal amount of polyA RNA was loaded on each well (Chester et al., 1998) and was prepared from the various organs as follows: (S) stomach; (LI) large intestine; (L) liver; (SP) spleen, (T) thymus; (K) kidney; (LG) lung; (C) cerebrum; (H) heart; (M) muscle; (MG) mammary gland; (O) ovary; (TE) testis; (SV) seminal vesicle. (E) RNA in-situ hybridization reveals that Zfp57 is expressed specifically in the maturing oocytes (purple stain). Frozen sections of the wild-type ovary were probed with an antisense riboprobe derived from a 0.5kb fragment of the 5’ portion of the Zfp57 cDNA. Arrows point to the labelled oocytes inside the cavity of the follicles.
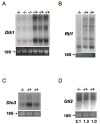
Figure 2. Loss of Zfp57 affects expression of the genes in the Dlk1-Dio3 imprinted region
(A) Total RNA samples from three homozygous (-/-) and three littermate heterozygous (-/+) embryos derived from the same null mother were probed with Dlk1 cDNA in Northern blot. (B), (C), (D) Equal amount of polyA RNA samples from a homozygous (-/-) and two littermate heterozygous (-/+) embryos derived from the same null mother were hybridized with a probe derived from Rtl1, Dio3 or Gtl2, respectively. Numbers in (D) indicate the band intensity of Gtl2 transcripts in each lane based on phosphor image analysis. Similar band intensity of 18S ribosomal RNA shown in the bottom panels of all figures indicates equal loading of the RNA samples. All embryos are in the pure 129 Sv/Ev background.
Figure 3. Loss of Zfp57 affects differential DNA methylation at the Dlk1-Dio3 and the Snrpn imprinted regions
Differential DNA methylation was assessed with the methylation sensitive restriction enzyme _HhaI. Stu_I-digested (Figure 3A) and _Pst_I–digested (Figure 3B) genomic DNA samples from embryos were HhaI digested and hybridized with a probe derived from the differentially methylated regions using Southern blot analysis (left panel). Histograms represent relative differential methylation levels based on quantitative phosphor image analysis of the band intensity of the southern blots shown on the left. The numbers above each bar indicate the representative lane on the Southern blot. Numbers within the bars indicate percentage of differential DNA methylation relative to that of wild-type embryos. Zero indicates absence of methylation. Genotypes and numbers analyzed for each class of embryo are indicated along the horizontal axis. All embryos are E11.5-E13 on a 129 Sv/Ev background. (A)The IG-DMR from the Dlk1-Dio3 region. Lane 1, wild-type embryos. Lane 2, heterozygous (+/-) embryos from a heterozygous (het) mother. Lane 3, homozygous (-/-) embryos from a het mother. Lane 4, heterozygous (-/+) embryos from a homozygous (null) mother. Lane 5, -/- embryos from a null mother. (B) The Snrpn DMR. Lane 1, wild-type embryos. Lane 2, +/- embryos from a het mother. Lane 3, -/- embryos from a het mother. Lane 4 and lane 5, -/+ embryos from a null mother. Lane 6, -/- embryos from a null mother and a het father. Lane 7, -/- embryos from a null mother and a null father. This Snrpn Southern data was further confirmed by COBRA analysis on samples including a subset analyzed by Southern blotting. A total of 25 mid-gestation heterozygous embryos from null mothers were assessed with 14 exhibiting methylation acquisition and 11 remaining unmethylated (data not shown).
Figure 4. Differential methylation at the Snrpn DMR is not established in absence of the maternal Zfp57 and can be acquired in the presence of the zygotic Zfp57
Genomic DNA samples from oocytes or embryos in the 129 Sv/Ev background were subjected to bisulphite sequencing. A total of 16 differentially methylated CpG sites at the Snrpn DMR are shown for Figures A-D. Filled oval, methylated CpG site. Open oval, unmethylated CpG sites. Line with ovals, a unique clone. A unique clone was assigned based on unconverted C residues in the sequences. The number of sequences is shown for a non-unique clone. The high conversion rate fails to distinguish whether these non-unique sequences represent clonal or individual products. A, Two different pools of unfertilized oocytes isolated from heterozygous female mice. B, Two different pools of unfertilized oocytes isolated from homozygous female mice. C, pooled E3.5 embryos. -/+ (E3.5), 29 heterozygous E3.5 embryos from the crosses between homozygous female mice and wild-type male mice. -/- (E3.5), 10 homozygous E3.5 embryos from the crosses between homozygous female mice and homozygous male mice. +/- (E3.5), 16 heterozygous E3.5 embryos from the crosses between wild-type female mice and homozygous male mice. D, genomic DNA samples were made from littermate E13.5 embryos from the cross between a homozygous female mouse and a heterozygous male mouse. E, Acquisition of DNA methylation imprint occurred on the maternally derived Snrpn DMR. Histograms are shown for the levels of methylation at the CpG sites of this DMR in E11.5-E13 embryos from the cross between a homozygous female mouse in the 129 Sv/Ev background and a wild-type male mouse in the DBA/2 background. Unfilled bars, eight littermate heterozygous embryos (1- 8) from this cross. Filled bar, seven littermate wild-type (WT) embryos from the cross between a wild-type 129 female mouse and a wild-type DBA/2 male mouse. Vertical axis, percentage of methylated CpG sites analyzed. Left panel, maternally derived 129 allele. Right panel, paternally derived DBA/2 allele.
Figure 5. Endogenous ZFP57 can bind to the Snrpn DMR region
A, Western blot analysis of affinity purified anti-ZFP57 polyclonal antibodies with total cell lysate samples. Lane 1, ZFP57 was over-expressed in COS cells. Lane 2, an ES clone containing one floxed allele (F) and one targeted allele (T) at the Zfp57 locus. Lane 3, wild-type TC1 ES cells. Lanes 4 and 5, two independent _Zfp57_-null ES clones carrying two deleted alleles. Please see Supplemental Figure S2 and Supplemental Data for the description of the targeted, floxed and deleted alleles. (B-E) Chromatin immunoprecipitation (ChIP) was performed with approximately one million ES cells. No ChIP PCR product was observed in the negative control samples without the addition of any antibodies (data not shown). B, the first round PCR product at the Snrpn DMR region. C, the second round PCR product at the Snrpn DMR region. D, the second round PCR product at a region about 40kb upstream of the Snrpn DMR region. E, the second round PCR product at the H19 DMR region. ChIP, ChIP PCR product. Input, PCR product from the input/starting samples; ns-IgG, ChIP PCR product from the control samples when rabbit non-specific IgG (ns-IgG) antibodies were added during immunoprecipitation. (B-E) Lane 1, an ES clone containing one floxed allele (F) and one targeted allele (T) at the Zfp57 locus. Lane 2, wild-type TC1 ES cells. Lanes 3 and 4, two independent _Zfp57_-null ES clones carrying two deleted alleles.
Figure 6. Maternal ZFP57 is present in pre-implantation embryos
Pre-implantation embryos were stained with affinity-purified polyclonal antibodies against ZFP57 (Figures A-H). They were also incubated with Hoechst dye to illuminate genomic DNA (Figures A’-H’). A homozygous E1.5 embryo (A or A’) and a homozygous E3.5 embryo (D or D’) were derived from the cross between homozygous female mice and homozygous male mice. Littermate E1.5 embryos (B and C) and E3.5 embryos (F and G) were derived from the crosses between heterozygous female mice and homozygous male mice. B (or G), a homozygote (-/-). C (or F), a heterozygote (+/-). The wild-type E3.5 embryo (E) was derived from the cross between a wild-type female mouse and a wild-type male mouse. The heterozygous (-/+) E3.5 embryo (H) was from the cross between a homozygous female mouse and a wild-type male mouse.
Comment in
- A KRAB domain zinc finger protein in imprinting and disease.
Hirasawa R, Feil R. Hirasawa R, et al. Dev Cell. 2008 Oct;15(4):487-8. doi: 10.1016/j.devcel.2008.09.006. Dev Cell. 2008. PMID: 18854130
Similar articles
- Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain.
Zuo X, Sheng J, Lau HT, McDonald CM, Andrade M, Cullen DE, Bell FT, Iacovino M, Kyba M, Xu G, Li X. Zuo X, et al. J Biol Chem. 2012 Jan 13;287(3):2107-18. doi: 10.1074/jbc.M111.322644. Epub 2011 Dec 5. J Biol Chem. 2012. PMID: 22144682 Free PMC article. - Extending the maternal-zygotic effect with genomic imprinting.
Li X. Li X. Mol Hum Reprod. 2010 Sep;16(9):695-703. doi: 10.1093/molehr/gaq028. Epub 2010 Apr 9. Mol Hum Reprod. 2010. PMID: 20382782 - Maternal and zygotic Zfp57 modulate NOTCH signaling in cardiac development.
Shamis Y, Cullen DE, Liu L, Yang G, Ng SF, Xiao L, Bell FT, Ray C, Takikawa S, Moskowitz IP, Cai CL, Yang X, Li X. Shamis Y, et al. Proc Natl Acad Sci U S A. 2015 Apr 21;112(16):E2020-9. doi: 10.1073/pnas.1415541112. Epub 2015 Apr 6. Proc Natl Acad Sci U S A. 2015. PMID: 25848000 Free PMC article. - ZFP57 and the Targeted Maintenance of Postfertilization Genomic Imprints.
Takahashi N, Gray D, Strogantsev R, Noon A, Delahaye C, Skarnes WC, Tate PH, Ferguson-Smith AC. Takahashi N, et al. Cold Spring Harb Symp Quant Biol. 2015;80:177-87. doi: 10.1101/sqb.2015.80.027466. Cold Spring Harb Symp Quant Biol. 2015. PMID: 27325708 Review. - Transient neonatal diabetes mellitus and hypomethylation at additional imprinted loci: novel ZFP57 mutation and review on the literature.
Touati A, Errea-Dorronsoro J, Nouri S, Halleb Y, Pereda A, Mahdhaoui N, Ghith A, Saad A, Perez de Nanclares G, H'mida Ben Brahim D. Touati A, et al. Acta Diabetol. 2019 Mar;56(3):301-307. doi: 10.1007/s00592-018-1239-3. Epub 2018 Oct 13. Acta Diabetol. 2019. PMID: 30315371 Review.
Cited by
- Environmental exposures influence multigenerational epigenetic transmission.
Klibaner-Schiff E, Simonin EM, Akdis CA, Cheong A, Johnson MM, Karagas MR, Kirsh S, Kline O, Mazumdar M, Oken E, Sampath V, Vogler N, Wang X, Nadeau KC. Klibaner-Schiff E, et al. Clin Epigenetics. 2024 Oct 17;16(1):145. doi: 10.1186/s13148-024-01762-3. Clin Epigenetics. 2024. PMID: 39420431 Free PMC article. Review. - Imprinting as Basis for Complex Evolutionary Novelties in Eutherians.
Schuff M, Strong AD, Welborn LK, Ziermann-Canabarro JM. Schuff M, et al. Biology (Basel). 2024 Aug 31;13(9):682. doi: 10.3390/biology13090682. Biology (Basel). 2024. PMID: 39336109 Free PMC article. Review. - CTCF barrier breaking by ZFP661 promotes protocadherin diversity in mammalian brains.
Jin J, Ralls S, Wu E, Wolf G, Sun MA, Springer DA, Cosby RL, Senft AD, Macfarlan TS. Jin J, et al. bioRxiv [Preprint]. 2023 May 8:2023.05.08.539838. doi: 10.1101/2023.05.08.539838. bioRxiv. 2023. PMID: 39185186 Free PMC article. Preprint. - Imprinted DNA methylation of the H19 ICR is established and maintained in vivo in the absence of Kaiso.
Matsuzaki H, Kimura M, Morihashi M, Tanimoto K. Matsuzaki H, et al. Epigenetics Chromatin. 2024 Jun 5;17(1):20. doi: 10.1186/s13072-024-00544-8. Epigenetics Chromatin. 2024. PMID: 38840164 Free PMC article. - The phenomenon of genomic imprinting was discovered 40 years ago.
Ferguson-Smith AC, Bartolomei MS. Ferguson-Smith AC, et al. Nature. 2024 May;629(8013):763-765. doi: 10.1038/d41586-024-01338-4. Nature. 2024. PMID: 38745025 No abstract available.
References
- Ahn JI, Lee KH, Shin DM, Shim JW, Lee JS, Chang SY, Lee YS, Brownstein MJ, Lee SH. Comprehensive transcriptome analysis of differentiation of embryonic stem cells into midbrain and hindbrain neurons. Dev Biol. 2004;265:491–501. - PubMed
- Akagi T, Usuda M, Matsuda T, Ko MS, Niwa H, Asano M, Koide H, Yokota T. Identification of Zfp-57 as a downstream molecule of STAT3 and Oct-3/4 in embryonic stem cells. Biochem Biophys Res Commun. 2005;331:23–30. - PubMed
- Arnaud P, Monk D, Hitchins M, Gordon E, Dean W, Beechey CV, Peters J, Craigen W, Preece M, Stanier P, et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Human molecular genetics. 2003;12:1005–1019. - PubMed
- Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, Tanaka K, Torigoe K, Rauscher FJ., 3rd Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes & development. 2003;17:1855–1869. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0701196/MRC_/Medical Research Council/United Kingdom
- P01 HD047675-01A17046/HD/NICHD NIH HHS/United States
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases