A quantitative comparison of sRNA-based and protein-based gene regulation - PubMed (original) (raw)
A quantitative comparison of sRNA-based and protein-based gene regulation
Pankaj Mehta et al. Mol Syst Biol. 2008.
Abstract
Small non-coding RNAs (sRNAs) have important functions as genetic regulators in prokaryotes. sRNAs act post-transcriptionally through complementary pairing with target mRNAs to regulate protein expression. We use a quantitative approach to compare and contrast sRNAs with conventional transcription factors (TFs) to better understand the advantages of each form of regulation. In particular, we calculate the steady-state behavior, noise properties, frequency-dependent gain (amplification), and dynamical response to large input signals of both forms of regulation. Although the mean steady-state behavior of sRNA-regulated proteins exhibits a distinctive tunable threshold linear behavior, our analysis shows that transcriptional bursting leads to significantly higher intrinsic noise in sRNA-based regulation than in TF-based regulation in a large range of expression levels and limits the ability of sRNAs to perform quantitative signaling. Nonetheless, we find that sRNAs are better than TFs at filtering noise in input signals. Additionally, we find that sRNAs allow cells to respond rapidly to large changes in input signals. These features suggest a 'niche' for sRNAs in allowing cells to transition quickly yet reliably between distinct states. This functional niche is consistent with the widespread appearance of sRNAs in stress response and quasi-developmental networks in prokaryotes.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
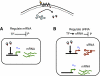
Genetic regulation through sRNAs. Left: small non-coding RNAs (sRNAs) regulate protein expression as part of a larger genetic network with a specific biological task (e.g. quorum sensing in Vibrio bacteria; Lenz et al, 2004). The sRNAs (stem loops) regulate target proteins by destabilizing target protein mRNAs (wavy lines), a stoichiometric process mediated by the RNA chaperone Hfq (hexagons). When the rate of sRNA transcription α_s_ greatly exceeds the rate of mRNA transcription α_m_, i.e. when, α_s_≫α_m_, nearly all the mRNAs are bound by sRNAs and cannot be translated. By contrast, when α_m_≫α_s_, there are many more mRNAs than sRNAs, and protein is highly produced. Right: the stochasticity (randomness) of cellular processes results in noise—statistical fluctuations in the molecular numbers. It is helpful to classify the total noise in the output (output noise) into (i) _input noise_—noise in the input signal from upstream components in the gene circuit, (ii) _intrinsic noise_—noise from stochasticity inherent in gene regulation through sRNAs, and (iii) _extrinsic noise_—all other sources of noise impinging on the signal processing system.
Figure 2
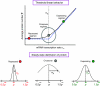
Steady-state behavior for gene regulation through sRNAs. For the regulated protein, the steady-state mean number p̄ exhibits an approximately threshold linear behavior as a function of the mRNA transcription rate α_m_. The threshold is set by the sRNA transcription rate α_s_. Protein expression can be classified into three regimens: repressed (α_s_≫α_m_), crossover (α_s_≈α_m_), and expressing (α_s_≪α_m_). In the repressed regimen, the average protein number is low. By contrast, the protein number increases almost linearly with α_m_ in the expressing regimen. The typical behavior of the noise α_p_, the standard deviation of the protein number, is shown for the three regimens.
Figure 3
Schematic drawing showing our comparison of transcriptional and post-transcriptional sRNA-mediated regulation. We take as the input signal to both systems a protein regulator (blue discs) that either directly transcriptionally regulates the relevant gene by acting as a repressor or transcriptionally regulates an sRNA acting as an activator. The protein regulator is chosen to have identical kinetic properties in both cases.
Figure 4
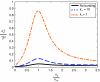
Protein noise with or without transcriptional bursting. Noise in protein expression σ_p_2/p̄_2 (variance divided by mean squared) as a function of the ratio of the sRNA and mRNA transcription rates, α_s/α_m_, for different levels of transcriptional bursting. We have assumed that both the sRNAs and mRNAs are produced in bursts. The noise peaks in the crossover regimen, α_s_≈α_m_. A slower unbinding rate _k_− for the repressor proteins controlling sRNA and mRNA expression results in larger transcriptional bursts. Parameters are (in min−1): α_m_=3, α_m_on=10, α_s_on=30, τ_m_=10, τ_s_=30, μ=0.02, α_p_=4, τ_p_=30, and k+ is adjusted to set the mean protein levels (for a discussion of parameter dependence, see Supplementary information).
Figure 5
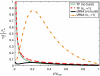
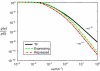
Comparison of analytic expressions for the intrinsic protein noise for TF- and sRNA-based regulation. The intrinsic noise for sRNA-based regulation as a function of normalized average protein concentration, p̄/_p_max, with and without transcriptional bursting, is shown. All parameters as in Figure 4.
Figure 6
Normalized frequency-dependent gain, g(ω)/g(0), as a function of the frequency, ω, for a small input signal for TF- and sRNA-based regulation in the repressed and expressing regimens. The amplitude of the frequency-dependent gain decreases rapidly ∝ω−4 at high frequencies for sRNAs compared with ∝ω−3 for TFs. Consequently, sRNA-based regulation is less sensitive to high-frequency input noise than TF-based regulation. Parameters as in Figure 4.
Figure 7
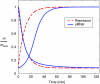
Large signal switching. Normalized mRNA level m/m_max, as a function of time, in response to step changes in the input, for both the sRNA- and TF-based regulation. Switching from high mRNA level (on state) to low mRNA level (off state) and vice versa. Switching from off to on state has a lag in the case of sRNA-based regulation, whereas the switching time from the on to off state for sRNAs is faster or comparable to that for TFs, depending on the choice of kinetic parameters. For sRNA-based regulation, α_m_=3.5 and α_s goes from ∼0.35 to 4.5 for switching from low to high and vice versa for high to low. For TF-based switching, α_m_ is such that both schemes have same steady states. Other parameters as in Figure 4.
Similar articles
- Quantitative characteristics of gene regulation by small RNA.
Levine E, Zhang Z, Kuhlman T, Hwa T. Levine E, et al. PLoS Biol. 2007 Sep;5(9):e229. doi: 10.1371/journal.pbio.0050229. PLoS Biol. 2007. PMID: 17713988 Free PMC article. - Direct comparison of small RNA and transcription factor signaling.
Hussein R, Lim HN. Hussein R, et al. Nucleic Acids Res. 2012 Aug;40(15):7269-79. doi: 10.1093/nar/gks439. Epub 2012 May 22. Nucleic Acids Res. 2012. PMID: 22618873 Free PMC article. - sRNA Target Prediction Organizing Tool (SPOT) Integrates Computational and Experimental Data To Facilitate Functional Characterization of Bacterial Small RNAs.
King AM, Vanderpool CK, Degnan PH. King AM, et al. mSphere. 2019 Jan 30;4(1):e00561-18. doi: 10.1128/mSphere.00561-18. mSphere. 2019. PMID: 30700509 Free PMC article. - Transcriptional and Post Transcriptional Control of Enterococcal Gene Regulation.
DebRoy S, Gao P, Garsin DA, Harvey BR, Kos V, Nes IF, Solheim M. DebRoy S, et al. 2014 Feb 12. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston: Massachusetts Eye and Ear Infirmary; 2014–. 2014 Feb 12. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston: Massachusetts Eye and Ear Infirmary; 2014–. PMID: 24649509 Free Books & Documents. Review. - Small RNA-mediated regulation in bacteria: A growing palette of diverse mechanisms.
Dutta T, Srivastava S. Dutta T, et al. Gene. 2018 May 20;656:60-72. doi: 10.1016/j.gene.2018.02.068. Epub 2018 Mar 1. Gene. 2018. PMID: 29501814 Review.
Cited by
- Measurement of the copy number of the master quorum-sensing regulator of a bacterial cell.
Teng SW, Wang Y, Tu KC, Long T, Mehta P, Wingreen NS, Bassler BL, Ong NP. Teng SW, et al. Biophys J. 2010 May 19;98(9):2024-31. doi: 10.1016/j.bpj.2010.01.031. Biophys J. 2010. PMID: 20441767 Free PMC article. - cis-antisense RNA, another level of gene regulation in bacteria.
Georg J, Hess WR. Georg J, et al. Microbiol Mol Biol Rev. 2011 Jun;75(2):286-300. doi: 10.1128/MMBR.00032-10. Microbiol Mol Biol Rev. 2011. PMID: 21646430 Free PMC article. Review. - Paradoxical suppression of small RNA activity at high Hfq concentrations due to random-order binding.
Sagawa S, Shin JE, Hussein R, Lim HN. Sagawa S, et al. Nucleic Acids Res. 2015 Sep 30;43(17):8502-15. doi: 10.1093/nar/gkv777. Epub 2015 Aug 10. Nucleic Acids Res. 2015. PMID: 26261213 Free PMC article. - Twins, quadruplexes, and more: functional aspects of native and engineered RNA self-assembly in vivo.
Lease RA, Arluison V, Lavelle C. Lease RA, et al. Front Life Sci. 2012 Mar;6(1-2):19-32. doi: 10.1080/21553769.2012.761163. Epub 2013 Mar 21. Front Life Sci. 2012. PMID: 23914307 Free PMC article. - The Tip of the Iceberg: On the Roles of Regulatory Small RNAs in the Virulence of Enterohemorrhagic and Enteropathogenic Escherichia coli.
Bhatt S, Egan M, Jenkins V, Muche S, El-Fenej J. Bhatt S, et al. Front Cell Infect Microbiol. 2016 Sep 21;6:105. doi: 10.3389/fcimb.2016.00105. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27709103 Free PMC article. Review.
References
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, Barkai N (2006) Noise in protein expression scales with natural protein abundance. Nat Genet 38: 636–643 - PubMed
- Berg HC (2003) E. coli in Motion. New York, USA: Springer-Verlag
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ (2006) Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell 24: 853–865 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K25 GM086909/GM/NIGMS NIH HHS/United States
- K25 GM086909-01/GM/NIGMS NIH HHS/United States
- P50 GM071508/GM/NIGMS NIH HHS/United States
- GM071508/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous