Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts - PubMed (original) (raw)
Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts
Jianghua Wang et al. Cancer Res. 2008.
Abstract
TMPRSS2/ERG gene fusions are found in the majority of prostate cancers; however, there is significant heterogeneity in the 5' region of the alternatively spliced fusion gene transcripts. We have found that there is also significant heterogeneity within the coding exons as well. There is variable inclusion of a 72-bp exon and other novel alternatively spliced isoforms. To assess the biological significance of these alternatively spliced transcripts, we expressed various transcripts in primary prostatic epithelial cells (PrEC) and in an immortalized PrEC line, PNT1a. The fusion gene transcripts promoted proliferation, invasion, and motility with variable activities that depended on the structure of the 5' region encoding the TMPRSS2/ERG fusion and the presence of the 72-bp exon. Cotransfection of different isoforms further enhanced biological activity, mimicking the situation in vivo, in which multiple isoforms are expressed. Finally, knockdown of the fusion gene in VCaP cells resulted in inhibition of proliferation in vitro and tumor progression in an in vivo orthotopic mice model. Our results indicate that TMPRSS2/ERG fusion isoforms have variable biological activities promoting tumor initiation and progression and are consistent with our previous clinical observations indicating that certain TMPRSS2/ERG fusion isoforms are significantly correlated with more aggressive disease.
Figures
Figure 1. Expression of a 72-bp exon in prostate cancer tissues expressing the TMPRSS2/ERG fusion gene
RT-PCR amplification of ERG alternatively spliced isoforms with or without 72-bp exon in using primers spanning this exon. A. Fusion gene expressing cancer tissues; B. Fusion negative cancer tissues; C. Benign tissues from the peripheral zone (14 tissues) or hyperplastic transition zone (6 tissues) which were free of cancer on pathological examination. D. VCaP cells.
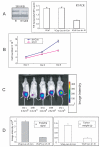
Figure 2. TMPRSS2/ERG fusion isoforms increase primary prostatic epithelial cell (PrEC) proliferation
A. Expression level of fusion gene in PrEC cells by real-time PCR, normalized to β-actin. B. Proliferation of the three groups of PrEC cells expressing TMPRSS2/ERG fusion Type III+72, (III+72) + (VI+72) or empty vector were measured using a Coulter counter. Cells (2.5 × 104) of each cell group were plated in 35mm dishes in complete medium. Cells were trypsinized and counted at day 3, 6 and 9 in triplicate. Mean +/− standard deviation is shown
Figure 3. TMPRSS2/ERG fusion isoforms affect PNT1a cell proliferation, invasion and motility
A. Expression level of fusion gene in PNT1a cells was evaluated by real-time PCR, normalized to β-actin. The growth PNT1a cells expressing TMPRSS2/ERG fusion Type III+72, VI+72, (III+72) + (VI+72) or empty vector was measured by using a Coulter counter. Cells (2.5 × 104) of each cell group were plated in 35mm dishes in complete medium. Cells were trypsinized and counted at day 3, 5 and 7 in triplicate. Mean +/− standard deviation is shown. B. Proliferation of 9 groups of PNT1a cells with overexpression of TMPRSS2/ERG isoforms individually or in combination and control cells. Cell numbers were counted at day 2, 4 and 6. C. Matrigel invasion of 9 groups of PNT1a cells with overexpression of TMPRSS2/ERG isoforms individually or in combination and control cells. Cells (2.5 × 104) of each group were plated into each well at day 0, and after 48h, the non-invading cells were removed from the upper surface of the membrane and the invading cells were stained and counted under the microscope. Assays were performed in triplicate. Mean +/− standard deviation is shown. D. PNT1a cells transfected with empty vector or TMPRSS2/ERG fusion types III, VI+72 and III+(VI+72) were seeded at 2.5×106 in 60-mm diameter culture dishes in complete medium. Cells were gently scraped with a plastic tip. The medium was removed, and cells were washed twice with PBS. Complete medium was added and cells were allowed to scatter/migrate into the area of clearing for a total of 72 hr and photomicrographs taken at 0h, 24h, 48h, and 72h time points. Scratch assays were performed four times and representative results are shown.
Figure 4. Homo- and heterodimerization of TMPRSS2/ERG fusion isoforms
A. M2 (Flag) tagged Type VI and VI+72 fusion genes or Hip1 protein were co-transfected with four fusion isoforms with a V5 tag in 293 cells, immunoprecipitated with anti-V5 antibody, and Western blot performed using anti-M2 antibody. Hip1 tagged with M2, cotransfected with Type VI+72 with a V5 tag was the negative control. B. Expression level of type VI, VI+72 and Hip1 proteins detected by anti-M2 antibody; C. Expression level of Type III, III+72, VI and VI+72 proteins detected by anti-V5 antibody; D. β-actin input control for each sample.
Figure 5. Knockdown of the TMPRSS2/ERG fusion in VCaP cells decreases cell proliferation in vitro and tumor progression in vivo
A. 293 cells were transiently transfected with V5-tagged Type III fusion gene expression construct and infected with lentivirus carrying shRNA against Type III fusion and protein expression evaluated using Western blotting with anti-V5 antibody. β-actin loading control is shown. Expression level of fusion gene mRNA in VCaP shRNA negative control or VCaP cells sh-III cells by real-time PCR normalized to β-actin. Mean +/− standard deviation. B. Cell growth curve comparing VCaP-Luc-sh-con and Vcap-Luc-sh-III cell numbers. Mean +/− standard deviation. C. Luciferase imaging of tumor growth in live mice from one cage by IVIS imaging system at 4 week time point. Three control (sh(−)) and two shRNA expressing (shIII) tumors are shown along with the corresponding image intensity. D. Luciferase imaging signal at 4 weeks after orthotopic injection and tumor weight at same time point.
Figure 6. Summary of reported ERG transcripts and identified ERG isoforms in prostate
A. The nine reported ERG isoforms are listed in left column with NCBI accession number and predicted protein weight; 17 reported exons are aligned on Chr.21 genomic sequence from 25364248 to 257322921, the size of each exon listed on the top of each exon. The 72-bp exon is highlighted by red color, the novel 61bp exon is shown in green. The 11 exons of ERG 2 are indicated. In-frame start codons are indicated by (+) and stop codons by (*). B. ERG isoforms in prostate cancer: arrows with different colors stand for different primers used to amplify the ERG transcripts. Sub-variants are indicated by letters.
Similar articles
- Role of the TMPRSS2-ERG gene fusion in prostate cancer.
Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, Mehra R, Chinnaiyan AM. Tomlins SA, et al. Neoplasia. 2008 Feb;10(2):177-88. doi: 10.1593/neo.07822. Neoplasia. 2008. PMID: 18283340 Free PMC article. - Quantitative analysis of ERG expression and its splice isoforms in formalin-fixed, paraffin-embedded prostate cancer samples: association with seminal vesicle invasion and biochemical recurrence.
Hagen RM, Adamo P, Karamat S, Oxley J, Aning JJ, Gillatt D, Persad R, Ladomery MR, Rhodes A. Hagen RM, et al. Am J Clin Pathol. 2014 Oct;142(4):533-40. doi: 10.1309/AJCPH88QHXARISUP. Am J Clin Pathol. 2014. PMID: 25239421 - Highly specific targeting of the TMPRSS2/ERG fusion gene using liposomal nanovectors.
Shao L, Tekedereli I, Wang J, Yuca E, Tsang S, Sood A, Lopez-Berestein G, Ozpolat B, Ittmann M. Shao L, et al. Clin Cancer Res. 2012 Dec 15;18(24):6648-57. doi: 10.1158/1078-0432.CCR-12-2715. Epub 2012 Oct 10. Clin Cancer Res. 2012. PMID: 23052253 Free PMC article. - 5' UTR control of native ERG and of Tmprss2:ERG variants activity in prostate cancer.
Zammarchi F, Boutsalis G, Cartegni L. Zammarchi F, et al. PLoS One. 2013;8(3):e49721. doi: 10.1371/journal.pone.0049721. Epub 2013 Mar 5. PLoS One. 2013. PMID: 23472063 Free PMC article. - [The progress of TMPRSS2-ETS gene fusions and their mechanism in prostate cancer].
Guo XQ, Gui YT, Cai ZM. Guo XQ, et al. Yi Chuan. 2011 Feb;33(2):117-22. doi: 10.3724/sp.j.1005.2011.00117. Yi Chuan. 2011. PMID: 21377967 Review. Chinese.
Cited by
- FGF23 promotes prostate cancer progression.
Feng S, Wang J, Zhang Y, Creighton CJ, Ittmann M. Feng S, et al. Oncotarget. 2015 Jul 10;6(19):17291-301. doi: 10.18632/oncotarget.4174. Oncotarget. 2015. PMID: 26019137 Free PMC article. - ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors.
Oh S, Shin S, Janknecht R. Oh S, et al. Biochim Biophys Acta. 2012 Aug;1826(1):1-12. doi: 10.1016/j.bbcan.2012.02.002. Epub 2012 Mar 8. Biochim Biophys Acta. 2012. PMID: 22425584 Free PMC article. Review. - The TMPRSS2-ERG Gene Fusion Blocks XRCC4-Mediated Nonhomologous End-Joining Repair and Radiosensitizes Prostate Cancer Cells to PARP Inhibition.
Chatterjee P, Choudhary GS, Alswillah T, Xiong X, Heston WD, Magi-Galluzzi C, Zhang J, Klein EA, Almasan A. Chatterjee P, et al. Mol Cancer Ther. 2015 Aug;14(8):1896-906. doi: 10.1158/1535-7163.MCT-14-0865. Epub 2015 May 29. Mol Cancer Ther. 2015. PMID: 26026052 Free PMC article. - Androgens Induce Functional CXCR4 through ERG Factor Expression in TMPRSS2-ERG Fusion-Positive Prostate Cancer Cells.
Cai J, Kandagatla P, Singareddy R, Kropinski A, Sheng S, Cher ML, Chinni SR. Cai J, et al. Transl Oncol. 2010 Jun 1;3(3):195-203. doi: 10.1593/tlo.09328. Transl Oncol. 2010. PMID: 20563261 Free PMC article. - FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data.
Sboner A, Habegger L, Pflueger D, Terry S, Chen DZ, Rozowsky JS, Tewari AK, Kitabayashi N, Moss BJ, Chee MS, Demichelis F, Rubin MA, Gerstein MB. Sboner A, et al. Genome Biol. 2010;11(10):R104. doi: 10.1186/gb-2010-11-10-r104. Epub 2010 Oct 21. Genome Biol. 2010. PMID: 20964841 Free PMC article.
References
- Mitelman F. Recurrent chromosome aberrations in cancer. Mutat Res. 2000;462:247–53. - PubMed
- Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. - PubMed
- Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–51. - PubMed
- Clark J, Merson S, Jhavar S, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26:2667–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical