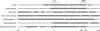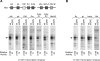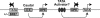Caudal, a key developmental regulator, is a DPE-specific transcriptional factor - PubMed (original) (raw)
Caudal, a key developmental regulator, is a DPE-specific transcriptional factor
Tamar Juven-Gershon et al. Genes Dev. 2008.
Abstract
The regulation of gene transcription is critical for the proper development and growth of an organism. The transcription of protein-coding genes initiates at the RNA polymerase II core promoter, which is a diverse module that can be controlled by many different elements such as the TATA box and downstream core promoter element (DPE). To understand the basis for core promoter diversity, we explored potential biological functions of the DPE. We found that nearly all of the Drosophila homeotic (Hox) gene promoters, which lack TATA-box elements, contain functionally important DPE motifs that are conserved from Drosophila melanogaster to Drosophila virilis. We then discovered that Caudal, a sequence-specific transcription factor and key regulator of the Hox gene network, activates transcription with a distinct preference for the DPE relative to the TATA box. The specificity of Caudal activation for the DPE is particularly striking when a BRE(u) core promoter motif is associated with the TATA box. These findings show that Caudal is a DPE-specific activator and exemplify how core promoter diversity can be used to establish complex regulatory networks.
Figures
Figure 1.
The core promoters of Drosophila Hox genes contain a conserved DPE motif. The core promoter sequences of the indicated D. melanogaster Hox genes are shown. We initially analyzed the sequence conservation of the promoters in eight different Drosophila species by using VISTA tools and CLUSTALW (Supplemental Fig. 1; data not shown). Based on this sequence alignment, positions that are identical between D. melanogaster and D. virilis (in terms of sequence and spacing relative to the A + 1 in the Inr) are shaded. D. melanogaster and D. virilis are estimated to be separated by an evolutionary period of ∼40–60 million years.
Figure 2.
The DPE is present in many genes that regulate the development of the Drosophila body plan. (A) Most of the Hox genes contain DPE-dependent core promoters. A schematic diagram of the Hox gene cluster is shown at the top. (lab) labial; (pb) proboscipedia; (Dfd) Deformed; (Scr) Sex combs reduced; (Antp P1) Antennapedia upstream promoter; (Antp P2) Antennapedia downstream promoter; (Ubx) Ultrabithorax; (abd-A) abdominal-A; (Abd-B) Abdominal-B. The wild-type (wt) and mutant DPE (mDPE) versions of the indicated core promoters (from −10 to +40 relative to the A + 1 start site) were subjected to in vitro transcription analysis with a Drosophila embryo nuclear extract. The resulting transcripts were detected by primer extension-reverse transcription analysis. (B) The core promoters of Caudal target genes contain functional DPE motifs. (ftz) fushi tarazu; (gt) giant; (h) hairy; (fkh) forkhead. In vitro transcription analysis was performed as in A.
Figure 3.
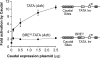
Caudal, a conserved master regulator of the Hox gene network, is a core-promoter-specific activator. (A) Caudal activates transcription with the Antp P2 DPE motif but not with the AdML TATA element. The two reporter constructs are identical except for the TATA and DPE sequences. Drosophila S2 cells were transfected with firefly luciferase reporter constructs as well as a Caudal expression plasmid, where indicated. To normalize for transfection efficiency, cells were cotransfected with a Pol III-Renilla luciferase control plasmid and assayed for dual luciferase activity. Error bars represent the SEM. (B) The Adh distal enhancer activates transcription with both the AdML TATA box and the Antp P2 DPE motif. The four reporter constructs are identical except for the presence or absence of the TATA, DPE, and Adh distal enhancer, as indicated. The Adh distal enhancer contains two binding sites for the Adf-1 transcription factor, which is present in S2 cells. Transfection assays were performed with the indicated constructs as in A. Error bars represent the SEM. (C) Caudal activates transcription with the E74B DPE motif and, to a lesser extent, the Adh proximal TATA element. Transfection assays were performed with the indicated constructs as in A. Error bars represent the SEM.
Figure 4.
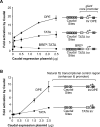
Transcriptional activation by Caudal is suppressed by the presence of a BREu motif upstream of the TATA box. The two reporter constructs are identical except for the absence or presence of the AdML BREu motif immediately upstream of the TATA box, and were analyzed as in Figure 3. Error bars represent the SEM.
Figure 5.
Caudal is a DPE-specific transcriptional activator. (A) Caudal preferentially activates transcription through the DPE in the gt core promoter. The reporter constructs contain six Caudal-binding sites upstream of the gt core promoter, which has both DPE and TATA motifs but lacks a BREu. The constructs are identical except for the mutation of the TATA and DPE motifs and the addition of the AdML BREu, where indicated. The transfection experiments were performed as in Figure 3. Error bars represent the SEM. (B) Caudal primarily activates through the DPE in the natural ftz transcriptional control region. The ftz core promoter contains both DPE and TATA motifs. The reporter constructs contain ftz enhancer and promoter sequences from −988 to +40 relative to the +1 start site, and are identical except for mutation of the DPE or TATA, as depicted. The transfection experiments were performed as in Figure 3. Error bars represent the SEM.
Figure 6.
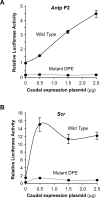
The natural Antp P2 and Scr promoters are activated by Caudal and are dependent on the DPE core promoter motif in S2 cells. (A) Antp P2 promoter. Constructs containing the wild-type and mutant versions of the Antp P2 promoter region (from −3033 to +61 relative to A + 1 in the Inr) were analyzed as in Figure 3. The activities are reported relative to the wild-type Antp P2 promoter in the absence of cotransfected Caudal expression plasmid, which was defined to be 1. Error bars represent the SEM. (B) Scr promoter. Constructs containing the wild-type and mutant versions of the Scr promoter region (from −3103 to +110 relative to A + 1 in the Inr) were analyzed as in Figure 3. The activities are reported relative to the wild-type Antp P2 promoter in the absence of cotransfected Caudal expression plasmid, which was defined to be 1. Error bars represent the SEM.
Figure 7.
A model for DPE-specific activation of transcription by Caudal. This model depicts a hypothetical segment of a regulatory network that comprises specific connections between activators and core promoter motifs. Caudal is a DPE-specific activator. TATA-specific activators might also exist. The ability of Caudal to function with a TATA box is further reduced by the presence of a BREu motif, which is located immediately upstream of a subset of TATA elements.
Similar articles
- Structure-Function Analysis of the Drosophila melanogaster Caudal Transcription Factor Provides Insights into Core Promoter-preferential Activation.
Shir-Shapira H, Sharabany J, Filderman M, Ideses D, Ovadia-Shochat A, Mannervik M, Juven-Gershon T. Shir-Shapira H, et al. J Biol Chem. 2015 Jul 10;290(28):17293-305. doi: 10.1074/jbc.M114.632109. Epub 2015 May 26. J Biol Chem. 2015. PMID: 26018075 Free PMC article. - Identification of evolutionarily conserved downstream core promoter elements required for the transcriptional regulation of Fushi tarazu target genes.
Shir-Shapira H, Sloutskin A, Adato O, Ovadia-Shochat A, Ideses D, Zehavi Y, Kassavetis G, Kadonaga JT, Unger R, Juven-Gershon T. Shir-Shapira H, et al. PLoS One. 2019 Apr 18;14(4):e0215695. doi: 10.1371/journal.pone.0215695. eCollection 2019. PLoS One. 2019. PMID: 30998799 Free PMC article. - The DPE, a core promoter element for transcription by RNA polymerase II.
Kadonaga JT. Kadonaga JT. Exp Mol Med. 2002 Sep 30;34(4):259-64. doi: 10.1038/emm.2002.36. Exp Mol Med. 2002. PMID: 12515390 Review. - Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters.
Burke TW, Kadonaga JT. Burke TW, et al. Genes Dev. 1996 Mar 15;10(6):711-24. doi: 10.1101/gad.10.6.711. Genes Dev. 1996. PMID: 8598298 - Regulation of gene expression via the core promoter and the basal transcriptional machinery.
Juven-Gershon T, Kadonaga JT. Juven-Gershon T, et al. Dev Biol. 2010 Mar 15;339(2):225-9. doi: 10.1016/j.ydbio.2009.08.009. Epub 2009 Aug 13. Dev Biol. 2010. PMID: 19682982 Free PMC article. Review.
Cited by
- From promoter motif to cardiac function: a single DPE motif affects transcription regulation and organ function in vivo.
Sloutskin A, Itzhak D, Vogler G, Pozeilov H, Ideses D, Alter H, Adato O, Shachar H, Doniger T, Shohat-Ophir G, Frasch M, Bodmer R, Duttke SH, Juven-Gershon T. Sloutskin A, et al. Development. 2024 Jul 15;151(14):dev202355. doi: 10.1242/dev.202355. Epub 2024 Jul 29. Development. 2024. PMID: 38958007 Free PMC article. - Onset of atonal expression in Drosophila retinal progenitors involves redundant and synergistic contributions of Ey/Pax6 and So binding sites within two distant enhancers.
Zhou Q, Zhang T, Jemc JC, Chen Y, Chen R, Rebay I, Pignoni F. Zhou Q, et al. Dev Biol. 2014 Feb 1;386(1):152-64. doi: 10.1016/j.ydbio.2013.11.012. Epub 2013 Nov 16. Dev Biol. 2014. PMID: 24247006 Free PMC article. - Dynamic modulation of enhancer responsiveness by core promoter elements in living Drosophila embryos.
Yokoshi M, Kawasaki K, Cambón M, Fukaya T. Yokoshi M, et al. Nucleic Acids Res. 2022 Jan 11;50(1):92-107. doi: 10.1093/nar/gkab1177. Nucleic Acids Res. 2022. PMID: 34897508 Free PMC article. - The RNA Polymerase II Core Promoter in Drosophila.
Vo Ngoc L, Kassavetis GA, Kadonaga JT. Vo Ngoc L, et al. Genetics. 2019 May;212(1):13-24. doi: 10.1534/genetics.119.302021. Genetics. 2019. PMID: 31053615 Free PMC article. Review. - Deduction and exploration of the evolution and function of vertebrate GFPT family.
Wei SA, Xu R, Ji YY, Ding ZW, Zou YZ. Wei SA, et al. Genes Genomics. 2022 Feb;44(2):175-185. doi: 10.1007/s13258-021-01188-8. Epub 2022 Jan 17. Genes Genomics. 2022. PMID: 35038160
References
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987;101:1–22. - PubMed
- Brooke N.M., Garcia-Fernàndez J., Holland P.W. The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature. 1998;392:920–922. - PubMed
- Burke T.W., Kadonaga J.T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & Dev. 1996;10:711–724. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases