Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knock-out cells - PubMed (original) (raw)
Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knock-out cells
Ya-Wen Liu et al. Mol Biol Cell. 2008 Dec.
Abstract
Dynamin (Dyn) is a multifunctional GTPase implicated in several cellular events, including endocytosis, intracellular trafficking, cell signaling, and cytokinesis. The mammalian genome encodes three isoforms, Dyn1, Dyn2, and Dyn3, and several splice variants of each, leading to the suggestion that distinct isoforms and/or distinct splice variants might mediate distinct cellular functions. We generated a conditional Dyn2 KO cell line and performed knockout and reconstitution experiments to explore the isoform- and splice variant specific cellular functions of ubiquitously expressed Dyn2. We find that Dyn2 is required for clathrin-mediated endocytosis (CME), p75 export from the Golgi, and PDGF-stimulated macropinocytosis and cytokinesis, but not for other endocytic pathways. Surprisingly, CME and p75 exocytosis were efficiently rescued by reintroduction of Dyn2, but not Dyn1, suggesting that these two isoforms function differentially in vesicular trafficking in nonneuronal cells. Both isoforms rescued macropinocytosis and cytokinesis, suggesting that dynamin function in these processes might be mechanistically distinct from its role in CME. Although all four Dyn2 splice variants could equally restore CME, Dyn2ba and -bb were more effective at restoring p75 exocytosis. This splice variant specificity correlated with their differential targeting to the Golgi. These studies reveal isoform and splice-variant specific functions for Dyn2.
Figures
Figure 1.
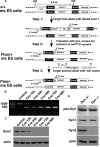
Generation of Dyn2 conditional KO cells. (A) Schematic diagram of the three-step strategy for generating Dyn2flox/− ES cells, by sequential transfection and selection of mouse ES cells with a conditional null vector and subsequent transfection and selection with a second null targeting vector. (B) PCR analysis of the Dyn2 KO cells after Cre adenovirus infection. DNA isolated at the indicated times after Cre adenovirus infection was used as a template to amplify the exon 1 region of Dyn2 to monitor excision efficiency (primer positions indicated in A). (C) Dyn2 protein depletion. Cell lysates were harvested at the indicated times after Cre adenovirus infection and Western-blotted with anti-Dyn2 or actin antibodies. (D) Dynamin expression in the Dyn2flox/− ES cells. Cell lysates from control or Cre adenovirus–infected cells were harvested 96 h after infection and blotted with anti-pan-dynamin or with the indicated isoform-specific antibodies.
Figure 2.
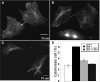
Cytokinesis defect in Dyn2 KO cells. Dyn2flox/− (A) or Dyn2 KO cells (B and C) were fixed and stained with anti-β-tubulin antibody to detect the midbody and the morphology of cells undergoing cytokinesis. Panels A and B are same scale, and C is a lower scale image to detect the long connection between two incompletely separated cells. (D) Quantitation of percentage of control, Dyn2 KO cells or Dyn2 KO cells reconstituted with either Dyn2 or -1, accumulating in late cytokinesis, based on detection of midbody staining. Error bars, SD of three independent experiments; 300 cells for each experiment were scored and counted.
Figure 3.
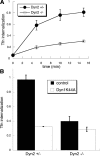
CME is potently inhibited in Dyn2 KO cells. (A) Kinetics of Tfn internalization in control and Dyn2 KO cells. Biotin-Tfn was incubated with Dyn2flox/− or Dyn2 KO cells for the indicated times at 37°C. Cells were returned to ice and internalized Tfn was detected as described in Materials and Methods. (B) Effects of Dyn1K44A overexpression on Tfn uptake in control and Dyn2 KO cells. Dyn2flox/− and Dyn2 KO cells were infected overnight with tTA-adenovirus alone (control) or together with Dyn1K44A-adenovirus. The extent of Tfn uptake after a 10-min incubation at 37°C was measured as described in Materials and Methods.
Figure 4.
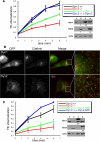
Dyn2 rescues CME in Dyn2 KO cells more efficiently than Dyn1. (A) Dyn2flox/− cells were infected with retroviruses encoding HA-Dyn1 (ba) or HA-Dyn2 (ba) and IRES-driven GFP, FACS-sorted for comparable levels of expression, and subsequently infected with Cre adenovirus to delete endogenous Dyn2. Their ability to internalize Tfn was assessed relative to control and Dyn2 KO cells as described above. The accompanying gels are Western blots with the indicated antibodies showing relative levels of expression (lane 1, Dyn2 KO cells; lane 2, Control cells; lane 3, Dyn2 KO cells reconstituted with HA-Dyn1; lane 4, Dyn2 KO cells reconstituted with HA-Dyn2). The small numbers below lanes 3 and 4 in B and C are fold-expression relative to endogenous dynamin after normalizing to actin loading controls. (B and C) Dyn2flox/− cells were infected with retroviruses encoding Dyn1 (ba)- or Dyn2 (ba)-GFP, FACS sorted for comparable levels of expression, and subsequently infected with Cre adenovirus to delete endogenous Dyn2. (B) Dyn2 KO cells expressing Dyn1-GFP or Dyn2-GFP and clathrin light chain-mCherry were fixed and observed under epi-fluorescence microscopy. (C) The kinetics of Tfn uptake in control, Dyn2 KO cells and Dyn2 KO cells expressing Dyn2-GFP or Dyn1-GFP as indicated. The accompanying gels are Western blots with the indicated antibodies showing relative levels of expression (lane 1, Dyn2 KO cells; lane 2, Control cells; lane 3, Dyn2 KO cells reconstituted with Dyn1-GFP; lane 4, Dyn2 KO cells reconstituted with Dyn2-GFP).
Figure 5.
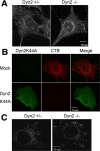
Dyn2 is not specifically required for cholera toxin, caveolae, or raft-mediated endocytosis. (A) Control and Dyn2 KO cells were incubated with Alexa fluor 594–conjugated cholera toxin B (CTB) for 5 min at 37°C. Surface-bound CTB was stripped by acid washing and internalized CTB detected by epifluorescence microscopy. (B) Dyn2flox/− cells were infected with Dyn2K44A or control adenovirus and then tested for CTB endocytosis as indicated in A. (C) BODIPY FL C5-LacSer uptake. Five-minute internalization of BODIPY FL C5-LacSer was analyzed in control and Dyn2 KO cells as described in Materials and Methods.
Figure 6.
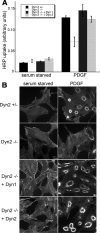
PDGF-stimulated macropinocytosis is supported by both Dyn1 and -2. (A) Fluid-phase uptake of HRP was measured in serum-starved cells before and after treatment with 10 ng/ml PDGF. Control and Dyn2 KO cells are equally effective at fluid-phase uptake under basal conditions, but Dyn2 KO cells are defective in PDGF-stimulated macropinocytosis. This defect is rescued by reexpression of either Dyn1 or -2. (B) Fluorescent phalloidin staining of actin in serum-starved and PDGF-treated cells reveals actin reorganization and dorsal ring formation upon PDGF stimulation. Dorsal ring formation is reduced in Dyn2 KO cells and restored by reintroduction of either Dyn1 or -2.
Figure 7.
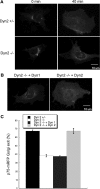
Dyn2 is specifically required for p75 export from the TGN. Control and Dyn2 KO cells were transfected with p75-mRFP/neurotropin receptor and at 24 h after infection, p75-mRFP was allowed to accumulate at the TGN as described in Materials and Methods. Cells were returned to 32°C to follow export of p75-mRFP from the TGN. (A) p75-mRFP accumulated in the TGN at t = 0 is largely depleted from the TGN at t = 40 min in control but not Dyn2 KO cells. (B) Reintroduction of Dyn2, but not Dyn1 can rescue p75-mRFP export in Dyn2 KO cells. (C) Quantification of p75-mRFP export in control, Dyn2 KO and Dyn1 or -2 reconstituted Dyn2 KO cells. The efficiency of p75-mRFP export from the TGN was determined by counting the percentage of cells without detectable p75-mRFP in the TGN. Three independent experiments and 100 cells for each experiment were scored and counted.
Figure 8.
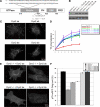
Dyn2 splice variants are specifically required for targeting and function at the Golgi. (A) Schematic and sequences of Dyn2 splice variants. (B) The HA-tagged Dyn2 aa, ab, ba, or bb splice variants were introduced into Dyn2 KO cells using IRES-GFP retrovirus vectors. Low expressing cells were selected by FACS sorting. Western blot using anti-Dyn2 antibody showing comparable expression levels of Dyn2 splice variants reintroduced into Dyn2 KO cells. (C) Localization of four Dyn2 splice variants. Four Dyn2 splice variants expressed in Dyn2 KO cells were detected using anti-HA antibody. (D) Tfn internalization was measured as described in Materials and Methods. All 4 Dyn2 splice variants function in CME. (E) Differential ability of Dyn2 splice variants to rescue p75-mRFP export from the TGN. (F) Quantification of the activity of different Dyn2 splice variants on p75-mRFP export. Three independent experiments as described in E were performed, and 100 cells from each experiment were scored for TGN exit of p75-mRFP.
Comment in
- Mol Biol Cell. 19:5031.
Similar articles
- A noncanonical role for dynamin-1 in regulating early stages of clathrin-mediated endocytosis in non-neuronal cells.
Srinivasan S, Burckhardt CJ, Bhave M, Chen Z, Chen PH, Wang X, Danuser G, Schmid SL. Srinivasan S, et al. PLoS Biol. 2018 Apr 18;16(4):e2005377. doi: 10.1371/journal.pbio.2005377. eCollection 2018 Apr. PLoS Biol. 2018. PMID: 29668686 Free PMC article. - Early and nonredundant functions of dynamin isoforms in clathrin-mediated endocytosis.
Bhave M, Mettlen M, Wang X, Schmid SL. Bhave M, et al. Mol Biol Cell. 2020 Aug 15;31(18):2035-2047. doi: 10.1091/mbc.E20-06-0363. Epub 2020 Jun 24. Mol Biol Cell. 2020. PMID: 32579424 Free PMC article. - Common membrane trafficking defects of disease-associated dynamin 2 mutations.
Liu YW, Lukiyanchuk V, Schmid SL. Liu YW, et al. Traffic. 2011 Nov;12(11):1620-33. doi: 10.1111/j.1600-0854.2011.01250.x. Epub 2011 Aug 5. Traffic. 2011. PMID: 21762456 Free PMC article. - Regulation of Clathrin-Mediated Endocytosis.
Mettlen M, Chen PH, Srinivasan S, Danuser G, Schmid SL. Mettlen M, et al. Annu Rev Biochem. 2018 Jun 20;87:871-896. doi: 10.1146/annurev-biochem-062917-012644. Epub 2018 Apr 16. Annu Rev Biochem. 2018. PMID: 29661000 Free PMC article. Review. - Reciprocal regulation of signaling and endocytosis: Implications for the evolving cancer cell.
Schmid SL. Schmid SL. J Cell Biol. 2017 Sep 4;216(9):2623-2632. doi: 10.1083/jcb.201705017. Epub 2017 Jul 3. J Cell Biol. 2017. PMID: 28674108 Free PMC article. Review.
Cited by
- A new role for the dynamin GTPase in the regulation of fusion pore expansion.
Anantharam A, Bittner MA, Aikman RL, Stuenkel EL, Schmid SL, Axelrod D, Holz RW. Anantharam A, et al. Mol Biol Cell. 2011 Jun 1;22(11):1907-18. doi: 10.1091/mbc.E11-02-0101. Epub 2011 Apr 1. Mol Biol Cell. 2011. PMID: 21460182 Free PMC article. - Insights into dynamin-associated disorders through analysis of equivalent mutations in the yeast dynamin Vps1.
Moustaq L, Smaczynska-de Rooij II, Palmer SE, Marklew CJ, Ayscough KR. Moustaq L, et al. Microb Cell. 2016 Mar 22;3(4):147-158. doi: 10.15698/mic2016.04.490. Microb Cell. 2016. PMID: 28357347 Free PMC article. - A nibbling mechanism for clathrin-mediated retrieval of secretory granule membrane after exocytosis.
Bittner MA, Aikman RL, Holz RW. Bittner MA, et al. J Biol Chem. 2013 Mar 29;288(13):9177-88. doi: 10.1074/jbc.M113.450361. Epub 2013 Feb 5. J Biol Chem. 2013. PMID: 23386611 Free PMC article. - SH3YL1 regulates dorsal ruffle formation by a novel phosphoinositide-binding domain.
Hasegawa J, Tokuda E, Tenno T, Tsujita K, Sawai H, Hiroaki H, Takenawa T, Itoh T. Hasegawa J, et al. J Cell Biol. 2011 May 30;193(5):901-16. doi: 10.1083/jcb.201012161. J Cell Biol. 2011. PMID: 21624956 Free PMC article. - Pharmacological inhibition of dynamin II reduces constitutive protein secretion from primary human macrophages.
Kockx M, Karunakaran D, Traini M, Xue J, Huang KY, Nawara D, Gaus K, Jessup W, Robinson PJ, Kritharides L. Kockx M, et al. PLoS One. 2014 Oct 27;9(10):e111186. doi: 10.1371/journal.pone.0111186. eCollection 2014. PLoS One. 2014. PMID: 25347775 Free PMC article.
References
- Albertson R., Riggs B., Sullivan W. Membrane traffic: a driving force in cytokinesis. Trends Cell Biol. 2005;15:92–101. - PubMed
- Antonescu C. N., Díaz M., Femia G., Planas J. V., Klip A. Clathrin-dependent and independent endocytosis of glucose transporter 4 (GLUT4) in myoblasts: regulation by mitochondrial uncoupling. Traffic. 2008;9:1173–1190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 MH061345/MH/NIMH NIH HHS/United States
- MH61345/MH/NIMH NIH HHS/United States
- R01 MH061345/MH/NIMH NIH HHS/United States
- GM42455/GM/NIGMS NIH HHS/United States
- R01 GM042455/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials