BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams - PubMed (original) (raw)
BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams
Tim Hulsen et al. BMC Genomics. 2008.
Abstract
Background: In many genomics projects, numerous lists containing biological identifiers are produced. Often it is useful to see the overlap between different lists, enabling researchers to quickly observe similarities and differences between the data sets they are analyzing. One of the most popular methods to visualize the overlap and differences between data sets is the Venn diagram: a diagram consisting of two or more circles in which each circle corresponds to a data set, and the overlap between the circles corresponds to the overlap between the data sets. Venn diagrams are especially useful when they are 'area-proportional' i.e. the sizes of the circles and the overlaps correspond to the sizes of the data sets. Currently there are no programs available that can create area-proportional Venn diagrams connected to a wide range of biological databases.
Results: We designed a web application named BioVenn to summarize the overlap between two or three lists of identifiers, using area-proportional Venn diagrams. The user only needs to input these lists of identifiers in the textboxes and push the submit button. Parameters like colors and text size can be adjusted easily through the web interface. The position of the text can be adjusted by 'drag-and-drop' principle. The output Venn diagram can be shown as an SVG or PNG image embedded in the web application, or as a standalone SVG or PNG image. The latter option is useful for batch queries. Besides the Venn diagram, BioVenn outputs lists of identifiers for each of the resulting subsets. If an identifier is recognized as belonging to one of the supported biological databases, the output is linked to that database. Finally, BioVenn can map Affymetrix and EntrezGene identifiers to Ensembl genes.
Conclusion: BioVenn is an easy-to-use web application to generate area-proportional Venn diagrams from lists of biological identifiers. It supports a wide range of identifiers from the most used biological databases currently available. Its implementation on the World Wide Web makes it available for use on any computer with internet connection, independent of operating system and without the need to install programs locally. BioVenn is freely accessible at http://www.cmbi.ru.nl/cdd/biovenn/.
Figures
Figure 1
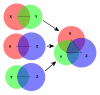
Construction of a three-circle BioVenn diagram. The method for generating a three-circle BioVenn diagram. The distance between the centers of each pair of circles is calculated, taking into account the size and overlap of the circles. Each pair of circles is put together using these distances. Then the three circles are put together, generating a three-circle diagram with not only two-circle overlaps but also a three-circle overlap.
Figure 2
The BioVenn input page. The BioVenn input page, with an example of three lists of Affymetrix probe identifiers.
Figure 3
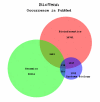
Example BioVenn diagram. The BioVenn diagram resulting from a PubMed comparison of the terms 'Bioinformatics', 'Genomics', and 'Systems Biology'.
Figure 4
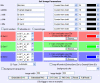
Current image statistics. The image statistics page belonging to the example from figure 3.
Similar articles
- Venn diagrams in bioinformatics.
Jia A, Xu L, Wang Y. Jia A, et al. Brief Bioinform. 2021 Sep 2;22(5):bbab108. doi: 10.1093/bib/bbab108. Brief Bioinform. 2021. PMID: 33839742 Review. - InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams.
Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. Heberle H, et al. BMC Bioinformatics. 2015 May 22;16(1):169. doi: 10.1186/s12859-015-0611-3. BMC Bioinformatics. 2015. PMID: 25994840 Free PMC article. - jvenn: an interactive Venn diagram viewer.
Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. Bardou P, et al. BMC Bioinformatics. 2014 Aug 29;15(1):293. doi: 10.1186/1471-2105-15-293. BMC Bioinformatics. 2014. PMID: 25176396 Free PMC article. - The Gene Set Builder: collation, curation, and distribution of sets of genes.
Yusuf D, Lim JS, Wasserman WW. Yusuf D, et al. BMC Bioinformatics. 2005 Dec 21;6:305. doi: 10.1186/1471-2105-6-305. BMC Bioinformatics. 2005. PMID: 16371163 Free PMC article. - DiVenn: An Interactive and Integrated Web-Based Visualization Tool for Comparing Gene Lists.
Sun L, Dong S, Ge Y, Fonseca JP, Robinson ZT, Mysore KS, Mehta P. Sun L, et al. Front Genet. 2019 May 3;10:421. doi: 10.3389/fgene.2019.00421. eCollection 2019. Front Genet. 2019. PMID: 31130993 Free PMC article. Review.
Cited by
- Nitroxoline resistance is associated with significant fitness loss and diminishes in vivo virulence of Escherichia coli.
Deschner F, Risch T, Baier C, Schlüter D, Herrmann J, Müller R. Deschner F, et al. Microbiol Spectr. 2024 Jan 11;12(1):e0307923. doi: 10.1128/spectrum.03079-23. Epub 2023 Dec 8. Microbiol Spectr. 2024. PMID: 38063385 Free PMC article. - MicroRNAs Expression in the Ileal Pouch of Patients with Ulcerative Colitis Is Robustly Up-Regulated and Correlates with Disease Phenotypes.
Ben-Shachar S, Yanai H, Sherman Horev H, Elad H, Baram L, Issakov O, Tulchinsky H, Pasmanik-Chor M, Shomron N, Dotan I. Ben-Shachar S, et al. PLoS One. 2016 Aug 18;11(8):e0159956. doi: 10.1371/journal.pone.0159956. eCollection 2016. PLoS One. 2016. PMID: 27536783 Free PMC article. - Bacterial Microcompartments Coupled with Extracellular Electron Transfer Drive the Anaerobic Utilization of Ethanolamine in Listeria monocytogenes.
Zeng Z, Boeren S, Bhandula V, Light SH, Smid EJ, Notebaart RA, Abee T. Zeng Z, et al. mSystems. 2021 Apr 13;6(2):e01349-20. doi: 10.1128/mSystems.01349-20. mSystems. 2021. PMID: 33850044 Free PMC article. - Proteomic analysis of the cyst stage of Entamoeba histolytica.
Ali IK, Haque R, Siddique A, Kabir M, Sherman NE, Gray SA, Cangelosi GA, Petri WA Jr. Ali IK, et al. PLoS Negl Trop Dis. 2012;6(5):e1643. doi: 10.1371/journal.pntd.0001643. Epub 2012 May 8. PLoS Negl Trop Dis. 2012. PMID: 22590659 Free PMC article. - High Mitochondrial Protein Expression as a Potential Predictor of Relapse Risk in Acute Myeloid Leukemia Patients with the Monocytic FAB Subtypes M4 and M5.
Selheim F, Aasebø E, Bruserud Ø, Hernandez-Valladares M. Selheim F, et al. Cancers (Basel). 2023 Dec 19;16(1):8. doi: 10.3390/cancers16010008. Cancers (Basel). 2023. PMID: 38201437 Free PMC article.
References
- Venn J. On the Diagrammatic and Mechanical Representation of Propositions and Reasonings. Philosophical Magazine and Journal of Science. 1880;9:1–18.
- Chow S, Ruskey F. Graph Drawing. Vol. 2912. Berlin/Heidelberg: Springer; 2004. Drawing Area-Proportional Venn and Euler Diagrams; pp. 466–477.
- VENNY. An interactive tool for comparing lists with Venn Diagrams http://bioinfogp.cnb.csic.es/tools/venny/
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases