The evolution of RNAi as a defence against viruses and transposable elements - PubMed (original) (raw)
Review
The evolution of RNAi as a defence against viruses and transposable elements
Darren J Obbard et al. Philos Trans R Soc Lond B Biol Sci. 2009.
Abstract
RNA interference (RNAi) is an important defence against viruses and transposable elements (TEs). RNAi not only protects against viruses by degrading viral RNA, but hosts and viruses can also use RNAi to manipulate each other's gene expression, and hosts can encode microRNAs that target viral sequences. In response, viruses have evolved a myriad of adaptations to suppress and evade RNAi. RNAi can also protect cells against TEs, both by degrading TE transcripts and by preventing TE expression through heterochromatin formation. The aim of our review is to summarize and evaluate the current data on the evolution of these RNAi defence mechanisms. To this end, we also extend a previous analysis of the evolution of genes of the RNAi pathways. Strikingly, we find that antiviral RNAi genes, anti-TE RNAi genes and viral suppressors of RNAi all evolve rapidly, suggestive of an evolutionary arms race between hosts and parasites. Over longer time scales, key RNAi genes are repeatedly duplicated or lost across the metazoan phylogeny, with important implications for RNAi as an immune defence.
Figures
Figure 1
RNAi pathways in Drosophila. (a) The antiviral siRNA pathway. Dicer-2 cuts dsRNA into siRNAs, which are loaded into an Argonaute-containing RISC that targets RNA for degradation. (b) The miRNA pathway. Primary miRNAs are transcribed from the genome, processed by Drosha and Dicer-1 into mature miRNAs. These are loaded into the Argonaute-containing effector complex (RISC), which binds mRNAs and recruits additional factors to inhibit translation. (c) The ‘ping-pong’ model of TE silencing in the Drosophila germ line. Aubergine and Argonaute-3 (Piwi family Argonautes) alternately cleave sense (red) and antisense (blue) transcripts from TEs, guided by piRNAs generated in the other half of the cycle. Cleavage both inactivates the transcript and generates the 5′ end of a new piRNA. The new piRNA-precursor is bound by the partner Piwi family member and the 3′ end degraded and then modified by the addition of a methyl group (Me). The nuclear localization of Piwi suggests that it might mediate heterochromatin assembly (Klattenhoff & Theurkauf 2008). It is unknown how this occurs, but one possibility is that the active Piwi complex binds nascent TE transcripts to recruit heterochromatin factors (Grewal & Elgin 2007). Recently, a fourth pathway has been identified, which targets TE transcripts in both the soma and the germ line, using Dcr-2 and Ago-2 from the antiviral pathway, but Loqs from the miRNA pathway (Chung et al. 2008; Czech et al. 2008).
Figure 2
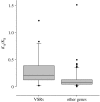
The evolutionary rate of VSRs in positive sense ssRNA plant viruses. The rate of protein evolution (_K_A, non-synonymous sites) relative to the rate of neutral evolution (_K_S, synonymous sites) for genes taken from 17 closely related pairs of ssRNA plant viruses. For viral suppressors of RNAi (VSRs) mean _K_A/_K_S=0.29 (_n_=20), and for other genes mean _K_A/_K_S=0.14 (_n_=72). In 14 of the 17 viruses, the rate of VSR evolution was higher than the average rate of the other genes (p<0.01, sign test). Where possible, genome pairs comprised two isolates of the same viral taxon (see the electronic supplementary material for accession details).
Figure 3
Evolutionary rate of RNAi-related genes in Drosophila. The rate of protein evolution (_K_A, non-synonymous sites) relative to the rate of neutral evolution (_K_S, synonymous sites) between D. melanogaster and D. simulans, plotted against aligned gene length. All 10 581 orthologous genes for which sequences are available are shown in grey, and 22 RNAi-pathway genes are shown as filled circles, squares and triangles. The smoothed median (red line) and the 75th, 85th and 95th percentiles (shaded background) are plotted for a sliding window (average 380 genes wide). RNAi-related genes evolve significantly more rapidly than the genome average (Wilcoxon signed-rank test, p<0.01). Six are in the fastest 5 per cent of genes (R2D2, Maelstrom, Krimper, Ago-2, Aubergine and Dcr-2), and a further 12 are in the upper 50 per cent. The piRNA pathway (purple circles) and viRNA pathway (red triangles) each evolve more rapidly than other genes (p<0.001 and p<0.01, respectively) but the miRNA pathway (green squares) does not (_p_=0.34). _K_A/KS was calculated by the method of Li et al. (1985). This analysis partially duplicates that of Obbard et al. (2006), but uses newer genome releases and an alternative estimator of _K_A/_K_S.
Figure 4
The rate of adaptive evolution in Drosophila RNAi genes. The estimated number of adaptive substitutions per codon (as opposed to neutral substitutions fixed by random genetic drift) for RNAi-pathway genes was estimated using the method of Smith & Eyre-Walker (2002). The analysis uses the D. simulans polymorphism data from Obbard et al. (; Ago-1, Ago-2, Dcr-1, Dcr-2, Loqs, R2D2) and Begun et al. (; all other genes), together with the fixed differences between these datasets and the D. melanogaster genome. Negative estimates (Ago-3 and Krimper) arise due to the relatively large number of amino acid polymorphisms in these genes. Genes with individually significant McDonald–Kreitman tests (McDonald & Kreitman 1991) are indicated by asterisks (*p<0.05, **p<0.01, ***p<0.001, no correction for multiple tests). Note that the differences in sample size mean that the power of the test varies greatly between genes (details available in the electronic supplementary material).
Figure 5
Evolutionary relationships of metazoans and their complements of selected RNAi genes. The presence of an RdRp and the number (where known) of SID and Dicer family members (Weinstock et al. 2006; Gordon & Waterhouse 2007) are indicated in table 1. The metazoan phylogeny is compiled from Bourlat et al. (2006), Delsuc et al. (2006) and Simionato et al. (2007). See figure 1 in the electronic supplementary material for sources.
Similar articles
- Metagenomic sequencing suggests a diversity of RNA interference-like responses to viruses across multicellular eukaryotes.
Waldron FM, Stone GN, Obbard DJ. Waldron FM, et al. PLoS Genet. 2018 Jul 30;14(7):e1007533. doi: 10.1371/journal.pgen.1007533. eCollection 2018 Jul. PLoS Genet. 2018. PMID: 30059538 Free PMC article. - RNA-Interference Pathways Display High Rates of Adaptive Protein Evolution in Multiple Invertebrates.
Palmer WH, Hadfield JD, Obbard DJ. Palmer WH, et al. Genetics. 2018 Apr;208(4):1585-1599. doi: 10.1534/genetics.117.300567. Epub 2018 Feb 1. Genetics. 2018. PMID: 29437826 Free PMC article. - RNAi: a defensive RNA-silencing against viruses and transposable elements.
Buchon N, Vaury C. Buchon N, et al. Heredity (Edinb). 2006 Feb;96(2):195-202. doi: 10.1038/sj.hdy.6800789. Heredity (Edinb). 2006. PMID: 16369574 Review. - Antiviral innate immune response of RNA interference.
Sidahmed A, Abdalla S, Mahmud S, Wilkie B. Sidahmed A, et al. J Infect Dev Ctries. 2014 Jul 14;8(7):804-10. doi: 10.3855/jidc.4187. J Infect Dev Ctries. 2014. PMID: 25022288 Review. - A DNA virus-encoded immune antagonist fully masks the potent antiviral activity of RNAi in Drosophila.
Bronkhorst AW, Vogels R, Overheul GJ, Pennings B, Gausson-Dorey V, Miesen P, van Rij RP. Bronkhorst AW, et al. Proc Natl Acad Sci U S A. 2019 Nov 26;116(48):24296-24302. doi: 10.1073/pnas.1909183116. Epub 2019 Nov 11. Proc Natl Acad Sci U S A. 2019. PMID: 31712431 Free PMC article.
Cited by
- The role of polymers in enabling RNAi-based technology for sustainable pest management.
Quilez-Molina AI, Niño Sanchez J, Merino D. Quilez-Molina AI, et al. Nat Commun. 2024 Oct 23;15(1):9158. doi: 10.1038/s41467-024-53468-y. Nat Commun. 2024. PMID: 39443470 Free PMC article. Review. - Neurodegenerative diseases reflect the reciprocal roles played by retroelements in regulating memory and immunity.
Herbert A. Herbert A. Front Neurosci. 2024 Sep 20;18:1445540. doi: 10.3389/fnins.2024.1445540. eCollection 2024. Front Neurosci. 2024. PMID: 39371608 Free PMC article. - Prevalent Fast Evolution of Genes Involved in Heterochromatin Functions.
Lin L, Huang Y, McIntyre J, Chang CH, Colmenares S, Lee YCG. Lin L, et al. Mol Biol Evol. 2024 Sep 4;41(9):msae181. doi: 10.1093/molbev/msae181. Mol Biol Evol. 2024. PMID: 39189646 Free PMC article. - The ancient Z-DNA and Z-RNA specific Zα fold has evolved modern roles in immunity and transcription through the natural selection of flipons.
Herbert A. Herbert A. R Soc Open Sci. 2024 Jun 19;11(6):240080. doi: 10.1098/rsos.240080. eCollection 2024 Jun. R Soc Open Sci. 2024. PMID: 39092141 Free PMC article. - Experimental Infection Models and Their Usefulness for White Spot Syndrome Virus (WSSV) Research in Shrimp.
Cox N, De Swaef E, Corteel M, Van Den Broeck W, Bossier P, Nauwynck HJ, Dantas-Lima JJ. Cox N, et al. Viruses. 2024 May 20;16(5):813. doi: 10.3390/v16050813. Viruses. 2024. PMID: 38793694 Free PMC article. Review.
References
- Ahlquist P. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science. 2002;296:1270–1273. doi:10.1126/science.1069132 - DOI - PubMed
- Alcami A., Koszinowski U.H. Viral mechanisms of immune evasion. Trends Microbiol. 2000;8:410–418. doi:10.1016/S0966-842X(00)01830-8 - DOI - PMC - PubMed
- Allen T.M., O'Connor D.H., Jing P.C., Dzuris J.L., Mothe B.R., Vogel T.U. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi:10.1038/35036559 - DOI - PubMed
- Anandalakshmi R., Pruss G.J., Ge X., Marathe R., Mallory A.C., Smith T.H. A viral suppressor of gene silencing in plants. Proc. Natl Acad. Sci. USA. 1998;95:13 079–13 084. doi:10.1073/pnas.95.22.13079 - DOI - PMC - PubMed
- Aravin A.A., Klenov M.S., Vagin V.V., Bantignies F., Cavalli G., Gvozdev V.A. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell. Biol. 2004;24:6742–6750. doi:10.1128/MCB.24.15.6742-6750.2004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases