Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms - PubMed (original) (raw)
Comparative Study
Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms
Peter A C 't Hoen et al. Nucleic Acids Res. 2008 Dec.
Abstract
The hippocampal expression profiles of wild-type mice and mice transgenic for deltaC-doublecortin-like kinase were compared with Solexa/Illumina deep sequencing technology and five different microarray platforms. With Illumina's digital gene expression assay, we obtained approximately 2.4 million sequence tags per sample, their abundance spanning four orders of magnitude. Results were highly reproducible, even across laboratories. With a dedicated Bayesian model, we found differential expression of 3179 transcripts with an estimated false-discovery rate of 8.5%. This is a much higher figure than found for microarrays. The overlap in differentially expressed transcripts found with deep sequencing and microarrays was most significant for Affymetrix. The changes in expression observed by deep sequencing were larger than observed by microarrays or quantitative PCR. Relevant processes such as calmodulin-dependent protein kinase activity and vesicle transport along microtubules were found affected by deep sequencing but not by microarrays. While undetectable by microarrays, antisense transcription was found for 51% of all genes and alternative polyadenylation for 47%. We conclude that deep sequencing provides a major advance in robustness, comparability and richness of expression profiling data and is expected to boost collaborative, comparative and integrative genomics studies.
Figures
Figure 1.
Categorization and abundance of tags. Distribution (in percentage of total) of unique tags (black bars) and individual reads (counts; open bars) over different categories (average from eight samples): high-confidence transcripts (canonical), low-confidence transcripts (noncanonical), mitochondrial RNA (mito), ribosomal RNA (ribo), genomic region with no evidence for transcription (just genome), repetitive genomic region (repeats) and tags with no hits in the genome.
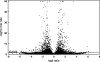
Figure 2.
Volcano plot of canonical tags. For every tag, the ratio in expression levels of transgenic over wild-type mice (2log scale, _x_-axis) is plotted against the Bayesian error rate (10log scale, _y_-axis). The horizontal line indicates the significance threshold applied, the 3179 differentially expressed tags being above that line. The plot shows that the tags with highest average differences between trasngenic and wild-type mice (far left and right part of the plot) are not all significant (due to large intragroup variation). The most significant tags (top of the plot) generally display small differences in expression between transgenic and wild-type but are, due to relatively high expression levels, very accurately measured and therefore display low intragroup variation.
Figure 3.
Correlation between absolute expression level (DGE) and microarrays signal intensity. Correlation of the tag abundance (square root transformed; _x_-axis) and intensities [normalized as described in (9)] on the five microarray platforms (_y_-axis) for matching ENSEMBL transcripts, for wild-type sample 1. Pearson correlations are indicated in the graphs. ABI: Applied Biosystems; AFF: Affymetrix; ILL: Illumina; AGL: Agilent; LGTC: home-spotted long oligonucleotide arrays.
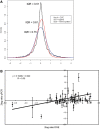
Figure 4.
Assessment of precision and accuracy of DGE. (A) Samples from the wild-type and transgenic pools were sequenced in three different lanes. We calculated the three possible independent log ratios between transgenic and wild-type samples (technical replicates). As a measure of precision, we determined the pair-wise differences between these technical replicates. The distribution of these differences is plotted as a density function (black line). This is also done for three technical replicates of wild-type over transgenic ratios determined on Agilent (red) and home-spotted (blue) microarrays. We balanced the number of observations per platform through random selection of 21 886 features. (B) As a measure of accuracy, we correlated logged ratios of the expression in transgenic versus wild-type mice as obtained by DGE (_x_-axis) against those obtained by qPCR (_y_-axis). All data and primer sequences can be found in
Supplementary Table 3
.
Similar articles
- Relative power and sample size analysis on gene expression profiling data.
van Iterson M, 't Hoen PA, Pedotti P, Hooiveld GJ, den Dunnen JT, van Ommen GJ, Boer JM, Menezes RX. van Iterson M, et al. BMC Genomics. 2009 Sep 17;10:439. doi: 10.1186/1471-2164-10-439. BMC Genomics. 2009. PMID: 19758461 Free PMC article. - 3' tag digital gene expression profiling of human brain and universal reference RNA using Illumina Genome Analyzer.
Asmann YW, Klee EW, Thompson EA, Perez EA, Middha S, Oberg AL, Therneau TM, Smith DI, Poland GA, Wieben ED, Kocher JP. Asmann YW, et al. BMC Genomics. 2009 Nov 16;10:531. doi: 10.1186/1471-2164-10-531. BMC Genomics. 2009. PMID: 19917133 Free PMC article. - Power of deep sequencing and agilent microarray for gene expression profiling study.
Feng L, Liu H, Liu Y, Lu Z, Guo G, Guo S, Zheng H, Gao Y, Cheng S, Wang J, Zhang K, Zhang Y. Feng L, et al. Mol Biotechnol. 2010 Jun;45(2):101-10. doi: 10.1007/s12033-010-9249-6. Mol Biotechnol. 2010. PMID: 20432071 - Improving reliability and performance of DNA microarrays.
Sievertzon M, Nilsson P, Lundeberg J. Sievertzon M, et al. Expert Rev Mol Diagn. 2006 May;6(3):481-92. doi: 10.1586/14737159.6.3.481. Expert Rev Mol Diagn. 2006. PMID: 16706748 Review. - RNA profiling in peripheral blood cells by fluorescent differential display PCR.
Steinau M, Rajeevan MS. Steinau M, et al. Methods Mol Biol. 2009;496:211-22. doi: 10.1007/978-1-59745-553-4_14. Methods Mol Biol. 2009. PMID: 18839113 Review.
Cited by
- Transcriptome Analysis of Beta macrocarpa and Identification of Differentially Expressed Transcripts in Response to Beet Necrotic Yellow Vein Virus Infection.
Fan H, Zhang Y, Sun H, Liu J, Wang Y, Wang X, Li D, Yu J, Han C. Fan H, et al. PLoS One. 2015 Jul 21;10(7):e0132277. doi: 10.1371/journal.pone.0132277. eCollection 2015. PLoS One. 2015. PMID: 26196682 Free PMC article. - Gene set enrichment analysis of RNA-Seq data: integrating differential expression and splicing.
Wang X, Cairns MJ. Wang X, et al. BMC Bioinformatics. 2013;14 Suppl 5(Suppl 5):S16. doi: 10.1186/1471-2105-14-S5-S16. Epub 2013 Apr 10. BMC Bioinformatics. 2013. PMID: 23734663 Free PMC article. - A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae.
Nookaew I, Papini M, Pornputtapong N, Scalcinati G, Fagerberg L, Uhlén M, Nielsen J. Nookaew I, et al. Nucleic Acids Res. 2012 Nov 1;40(20):10084-97. doi: 10.1093/nar/gks804. Epub 2012 Sep 10. Nucleic Acids Res. 2012. PMID: 22965124 Free PMC article. - Comparative transcriptional profiling provides insights into the evolution and development of the zygomorphic flower of Vicia sativa (Papilionoideae).
Liu Z, Ma L, Nan Z, Wang Y. Liu Z, et al. PLoS One. 2013;8(2):e57338. doi: 10.1371/journal.pone.0057338. Epub 2013 Feb 21. PLoS One. 2013. PMID: 23437373 Free PMC article. - Increased sensitivity of next generation sequencing-based expression profiling after globin reduction in human blood RNA.
Mastrokolias A, den Dunnen JT, van Ommen GB, 't Hoen PA, van Roon-Mom WM. Mastrokolias A, et al. BMC Genomics. 2012 Jan 18;13:28. doi: 10.1186/1471-2164-13-28. BMC Genomics. 2012. PMID: 22257641 Free PMC article.
References
- Irizarry RA, Warren D, Spencer F, Kim IF, Biswal S, Frank BC, Gabrielson E, Garcia JG, Geoghegan J, Germino G, et al. Multiple-laboratory comparison of microarray platforms. Nat. Methods. 2005;2:345–350. - PubMed
- Harbers M, Carninci P. Tag-based approaches for transcriptome research and genome annotation. Nat. Methods. 2005;2:495–502. - PubMed
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000;18:630–634. - PubMed
- Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous