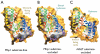Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter - PubMed (original) (raw)
. 2008 Oct 31;322(5902):709-13.
doi: 10.1126/science.1164440. Epub 2008 Oct 16.
Tatsuro Shimamura, Shunsuke Yajima, Shun'ichi Suzuki, Osman Mirza, Kuakarun Krusong, Elisabeth P Carpenter, Nicholas G Rutherford, Jonathan M Hadden, John O'Reilly, Pikyee Ma, Massoud Saidijam, Simon G Patching, Ryan J Hope, Halina T Norbertczak, Peter C J Roach, So Iwata, Peter J F Henderson, Alexander D Cameron
Affiliations
- PMID: 18927357
- PMCID: PMC2885439
- DOI: 10.1126/science.1164440
Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter
Simone Weyand et al. Science. 2008.
Abstract
The nucleobase-cation-symport-1 (NCS1) transporters are essential components of salvage pathways for nucleobases and related metabolites. Here, we report the 2.85-angstrom resolution structure of the NCS1 benzyl-hydantoin transporter, Mhp1, from Microbacterium liquefaciens. Mhp1 contains 12 transmembrane helices, 10 of which are arranged in two inverted repeats of five helices. The structures of the outward-facing open and substrate-bound occluded conformations were solved, showing how the outward-facing cavity closes upon binding of substrate. Comparisons with the leucine transporter LeuT(Aa) and the galactose transporter vSGLT reveal that the outward- and inward-facing cavities are symmetrically arranged on opposite sides of the membrane. The reciprocal opening and closing of these cavities is synchronized by the inverted repeat helices 3 and 8, providing the structural basis of the alternating access model for membrane transport.
Figures
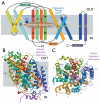
Fig. 1. Structure of Mhp1 from Microbacterum liquefaciens
(A) Mhp1 topology. The positions of the substrate and the cation binding sites are depicted as a brown ellipsoid and blue circle, respectively. The membrane is shown in grey and the outward-facing cavity observed in the structure is highlighted in light blue. The horizontal helices on the inside and outside of membrane are indicated as IN and OUT. TMs 3 and 8 pack onto each other in three dimensional space. (B) The Mhp1 structure viewed in the plane of the membrane. The figure is based on the high-resolution structure of the Mhp1 without benzyl-hydantoin. The position of the substrate in the Mhp1- benzyl-hydantoin complex structure is shown as a reference. A sodium ion is also shown and labelled. (C) View from the outside of the membrane.
Fig.2. Outward- and inward- facing cavities
(A) Slice through the surface of the Mhp1 substrate-free structure, viewed parallel to the membrane, showing the outward-facing cavity. The Connolly surface of Mph1 is shown in yellow (calculated with a probe radius of 2Å). The ribbon representation of Mhp1 is coloured as in Fig. 1. (B) As for panel A but for the Mhp1 substrate-occluded structure. Bound benzyl-hydantoin is shown in cyan. Note that the outward-facing cavity shown in panel A is blocked and the substrate is occluded from the outside of the membrane. (C) As for A, but for the vSGLT substrate-occluded structure (PDB accession code: 3DH4), showing the inward-facing cavity. Bound galactose is shown in cyan and red.
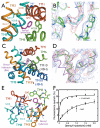
Fig. 3. Substrate and cation binding sites
(A) Substrate binding site. The helices and key interacting residues of the site are shown. The benzyl-hydantoin and the nearby sodium are shown in magenta and blue, respectively. (B) Electron density associated with benzyl-hydantoin. The 2Fo-Fc map (blue) has been contoured at 0.6σ and the Fo-Fc map (red) at 3 σ (C) The conformational change upon the substrate binding. Transmembrane helix (TM) 10 and the loop between TMs 9 and 10 show a conformational change that occludes the bound substrate from the outside of the membrane. The trace of this region, shown in grey, is superimposed on the substrate-free structure. Benzyl-hydantoin is abbreviated as BH in the figure. D) Electron density and model for TM 10. The 2Fo-Fc map (blue) has been contoured at 0.6σ and the Fo-Fc map (red) at 2 σ. The trace for the benzyl-hydantoin free structure is shown in magenta and that for the benzyl-hydantoin complex is depicted in green. The grey trace is for the vSGLT structure (PDB accession code: 3DH4) superimposed onto Mhp1. Note that the maps in B and D were calculated based on the molecular replacement solution using the substrate-free structure with residues 355-370 omitted so is not biased by these residues. See Materials and Methods (24) for details. (E) Helices and key residues surrounding the cation and substrate binding sites. (F) Tryptophan-fluorescence-quenching by benzyl-hydantoin. The Mhp1 solution was titrated by benzyl-hydantoin and the decrease in the tryptophan fluorescence at 348nm was monitored. The measurements were performed with (triangles) and without (circles) 15 mM NaCl in the solution. For details, see Materials and Methods (25).
Fig. 4
Proposed substrate translocation mechanism by Mhp1. Schematic diagram showing four different conformational states, namely “Outward-facing open”, “Outward-facing occluded”, “Inward-facing occluded” and “Inward-facing open”, are shown. Substrate and sodium binding sites are labelled as “S” and “Na+”, respectively.
Similar articles
- Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1.
Shimamura T, Weyand S, Beckstein O, Rutherford NG, Hadden JM, Sharples D, Sansom MS, Iwata S, Henderson PJ, Cameron AD. Shimamura T, et al. Science. 2010 Apr 23;328(5977):470-3. doi: 10.1126/science.1186303. Science. 2010. PMID: 20413494 Free PMC article. - Conformational cycle and ion-coupling mechanism of the Na+/hydantoin transporter Mhp1.
Kazmier K, Sharma S, Islam SM, Roux B, Mchaourab HS. Kazmier K, et al. Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):14752-7. doi: 10.1073/pnas.1410431111. Epub 2014 Sep 29. Proc Natl Acad Sci U S A. 2014. PMID: 25267652 Free PMC article. - The alternating access mechanism of transport as observed in the sodium-hydantoin transporter Mhp1.
Weyand S, Shimamura T, Beckstein O, Sansom MS, Iwata S, Henderson PJ, Cameron AD. Weyand S, et al. J Synchrotron Radiat. 2011 Jan;18(1):20-3. doi: 10.1107/S0909049510032449. Epub 2010 Nov 5. J Synchrotron Radiat. 2011. PMID: 21169684 Free PMC article. - Lessons from lactose permease.
Guan L, Kaback HR. Guan L, et al. Annu Rev Biophys Biomol Struct. 2006;35:67-91. doi: 10.1146/annurev.biophys.35.040405.102005. Annu Rev Biophys Biomol Struct. 2006. PMID: 16689628 Free PMC article. Review.
Cited by
- Structure and mechanism of a glutamate-GABA antiporter.
Ma D, Lu P, Yan C, Fan C, Yin P, Wang J, Shi Y. Ma D, et al. Nature. 2012 Mar 11;483(7391):632-6. doi: 10.1038/nature10917. Nature. 2012. PMID: 22407317 - Transmembrane domain 6 of the human serotonin transporter contributes to an aqueously accessible binding pocket for serotonin and the psychostimulant 3,4-methylene dioxymethamphetamine.
Field JR, Henry LK, Blakely RD. Field JR, et al. J Biol Chem. 2010 Apr 9;285(15):11270-80. doi: 10.1074/jbc.M109.093658. Epub 2010 Feb 16. J Biol Chem. 2010. PMID: 20159976 Free PMC article. - Mechanism of anion selectivity and stoichiometry of the Na+/I- symporter (NIS).
Paroder-Belenitsky M, Maestas MJ, Dohán O, Nicola JP, Reyna-Neyra A, Follenzi A, Dadachova E, Eskandari S, Amzel LM, Carrasco N. Paroder-Belenitsky M, et al. Proc Natl Acad Sci U S A. 2011 Nov 1;108(44):17933-8. doi: 10.1073/pnas.1108278108. Epub 2011 Oct 19. Proc Natl Acad Sci U S A. 2011. PMID: 22011571 Free PMC article. - Coupled ion binding and structural transitions along the transport cycle of glutamate transporters.
Verdon G, Oh S, Serio RN, Boudker O. Verdon G, et al. Elife. 2014 May 19;3:e02283. doi: 10.7554/eLife.02283. Elife. 2014. PMID: 24842876 Free PMC article.
References
- Jardetzky O. Nature. 1966;211:969. - PubMed
- Kaback HR, et al. Proc. Natl. Acad. Sci. USA. 2007;104:491. - PubMed
- Toyoshima C. Arch. Biochem. Biophys. 2008;476:3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/C51725X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/C51725/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G020043/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 062164/Z/00/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- B17935/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 079209/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources