Blm10 binds to pre-activated proteasome core particles with open gate conformation - PubMed (original) (raw)
Blm10 binds to pre-activated proteasome core particles with open gate conformation
Andrea Lehmann et al. EMBO Rep. 2008 Dec.
Erratum in
- EMBO Rep. 2009 Jan;10(1):101
Abstract
Blm10 is bound to the yeast proteasome core particle, a crucial protease of eukaryotic cells [corrected]. Two gates, at both ends of the CP, control the access of protein substrates to the catalytic cavity of the CP. Normally, substrate access is auto-inhibited by a closed gate conformation unless regulatory complexes are bound to the CP and translocate protein substrates in an ATP-dependent manner. Here, we provide evidence that Blm10 recognizes pre-activated open gate CPs, which are assumed to exist in an equilibrium with inactive closed gate CP. Consequently, single-capped Blm10-CP shows peptide hydrolysis activity. Under conditions of disturbed CP assembly, as well as in open gate mutants, pre-activated CP or constitutively active CP, respectively, prevail. Then, Blm10 sequesters disordered and open gate CP by forming double-capped Blm10(2)-CP in which peptide hydrolysis activity is repressed. We conclude that Blm10 distinguishes between gate conformations and regulates the activation of CP.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
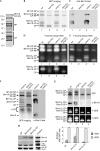
Pre-activated core particles are stabilized by Blm10 in _ump1_Δ cells. (A) Affinity-purified wild-type proteasomes (Lehmann et al, 2002) contain Blm10 and Ecm29, as identified by finger print mass spectroscopy; RP and CP subunits are embraced. (B–E) GFP-labelled CPs of isogenic wild-type (wt; lane 1), _blm10_Δ (lane 2), _ump1_Δ (lane 3) and _blm10_Δ _ump1_Δ cells (lane 4) were resolved by native PAGE; CP configurations are assigned. (B) CP distributions were visualized by GFP imaging. (C) Blm10-associated CPs were detected by Western blot. (D) By using Suc-LLVY-AMC (Y) as a peptide substrate, in-gel CP activity was detected in the absence and (E) in the presence of 0.02% SDS. The lower part of (D) is shown with high contrast. (F) Blm10 is neither associated with half-CP (Fehlker et al, 2003) nor completely recruited to CP. To visualize half-CP by GFP imaging, the contrast below the dotted line is increased. Blm10-associated CPs are detected by immunoblot. (G) GFP-labelled CP were affinity-purified from wild-type (lane 1), _ump1_Δ (lane 2), _blm10_Δ (lane 3) and _blm10_Δ _ump1_Δ (lane 4) cells, resolved by native PAGE, visualized by GFP imaging, probed for Blm10, and assayed for Y-activity in the absence and presence of SDS. Owing to the strong stimulation of CP activity by 0.02% SDS, the exposure time of the in-gel activity assays had to be reduced. The histogram shows arbitrary AMC/GFP ratios of Blm102-CP, Blm10-CP and CP. (H) Blm102-CP, Blm10-CP and CP of _ump1_Δ cells were extracted from the native gel, subjected to SDS–PAGE and analysed by Western blot as indicated; lysate acted as a control. Pac1 to Pac4 were not identified in mature CP (data not shown). AMC, 7-amino-4-methyl-coumarin; CP, core particle; GFP, green fluorescent protein; RP, regulatory particle; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.
Figure 2
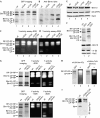
Constitutively active core particles are capped by Blm10 or RP. (A) Extracts of wild-type (wt; lane 1), α3ΔN (lane 2), α7ΔN (lane 3) and α3ΔN/α7ΔN (lane 4) cells were analysed by native PAGE and GFP imaging. Equal amounts of protein were loaded. (B) Blm10-bound CPs were identified by Western blot. (C) In-gel assays for Y-activity were performed in the absence of SDS or (D) in the presence of SDS. (E) Blm10 and β5 levels of wild type (lane 1), α3ΔN (lane 2), α7ΔN (lane 3) and α3ΔN/α7ΔN (lane 4) were determined by Western blot. (F) Extracts of wild-type cells (lane 1) and α4ΔN mutants (lane 2) expressing GFP-tagged α4 were analysed by native PAGE and GFP imaging. Blm10 and β5 levels were determined by Western blot. (G) Extracts of _pac1_Δ _pac2_Δ (upper panels), _pac3_Δ _pac4_Δ (lower panels) and isogenic wild-type cells, respectively, were analysed by native PAGE and GFP imaging. Y-activity assays were performed in the absence and presence of SDS. To visualize Blm10-CP activity, the upper in-gel activity assay was overexposed. Equal amounts of protein were loaded. (H) Native PAGE of α4ΔN _blm10_Δ cell extracts (GFP-tagged α4) were analysed by GFP imaging (lane 1) and Y-activity assay (lane 2). Native PAGE of α3ΔN/α7ΔN _blm10_Δ cell extracts (GFP-tagged β5) were analysed by GFP imaging (lane 3) and Y-activity assay (lane 4). (I) Extracts from wild-type (lane 1) and α3ΔN/α7ΔN _blm10_Δ (lane 2) cells were subjected to native PAGE, blotted and probed for Rpt1 (RP base). CP, core particle; GFP, green fluorescent protein; PAGE, polyacrylamide gel electrophoresis; RP, regulatory particle.
Figure 3
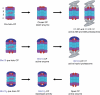
Model of Blm10 function. For a description of the model, see the Speculation section. CP, core particle; RP, regulatory particle.
Similar articles
- Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening.
Sadre-Bazzaz K, Whitby FG, Robinson H, Formosa T, Hill CP. Sadre-Bazzaz K, et al. Mol Cell. 2010 Mar 12;37(5):728-35. doi: 10.1016/j.molcel.2010.02.002. Mol Cell. 2010. PMID: 20227375 Free PMC article. - Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism.
Dange T, Smith D, Noy T, Rommel PC, Jurzitza L, Cordero RJ, Legendre A, Finley D, Goldberg AL, Schmidt M. Dange T, et al. J Biol Chem. 2011 Dec 16;286(50):42830-9. doi: 10.1074/jbc.M111.300178. Epub 2011 Oct 24. J Biol Chem. 2011. PMID: 22025621 Free PMC article. - The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle.
Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, Finley D. Schmidt M, et al. Nat Struct Mol Biol. 2005 Apr;12(4):294-303. doi: 10.1038/nsmb914. Epub 2005 Mar 20. Nat Struct Mol Biol. 2005. PMID: 15778719 - Proteasome activator 200: the heat is on..
Savulescu AF, Glickman MH. Savulescu AF, et al. Mol Cell Proteomics. 2011 May;10(5):R110.006890. doi: 10.1074/mcp.R110.006890. Epub 2011 Mar 9. Mol Cell Proteomics. 2011. PMID: 21389348 Free PMC article. Review. - Assembly and function of the proteasome.
Saeki Y, Tanaka K. Saeki Y, et al. Methods Mol Biol. 2012;832:315-37. doi: 10.1007/978-1-61779-474-2_22. Methods Mol Biol. 2012. PMID: 22350895 Review.
Cited by
- Dynamic Regulation of the 26S Proteasome: From Synthesis to Degradation.
Marshall RS, Vierstra RD. Marshall RS, et al. Front Mol Biosci. 2019 Jun 7;6:40. doi: 10.3389/fmolb.2019.00040. eCollection 2019. Front Mol Biosci. 2019. PMID: 31231659 Free PMC article. Review. - Nuclear Transport of Yeast Proteasomes.
Wendler P, Enenkel C. Wendler P, et al. Front Mol Biosci. 2019 May 16;6:34. doi: 10.3389/fmolb.2019.00034. eCollection 2019. Front Mol Biosci. 2019. PMID: 31157235 Free PMC article. Review. - Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures.
Guan H, Wang Y, Yu T, Huang Y, Li M, Saeed AFUH, Perčulija V, Li D, Xiao J, Wang D, Zhu P, Ouyang S. Guan H, et al. PLoS Biol. 2020 Mar 5;18(3):e3000654. doi: 10.1371/journal.pbio.3000654. eCollection 2020 Mar. PLoS Biol. 2020. PMID: 32134919 Free PMC article. - Structural roles of Ump1 and β-subunit propeptides in proteasome biogenesis.
Mark E, Ramos PC, Kayser F, Höckendorff J, Dohmen RJ, Wendler P. Mark E, et al. Life Sci Alliance. 2024 Sep 11;7(11):e202402865. doi: 10.26508/lsa.202402865. Print 2024 Nov. Life Sci Alliance. 2024. PMID: 39260885 Free PMC article. - Cationic porphyrins are tunable gatekeepers of the 20S proteasome.
Santoro AM, Cunsolo A, D'Urso A, Sbardella D, Tundo GR, Ciaccio C, Coletta M, Diana D, Fattorusso R, Persico M, Di Dato A, Fattorusso C, Milardi D, Purrello R. Santoro AM, et al. Chem Sci. 2016 Feb 1;7(2):1286-1297. doi: 10.1039/c5sc03312h. Epub 2015 Nov 9. Chem Sci. 2016. PMID: 29910886 Free PMC article.
References
- Baumeister W, Walz J, Zuhl F, Seemuller E (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell 92: 367–380 - PubMed
- Chen P, Hochstrasser M (1996) Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell 86: 961–972 - PubMed
- Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D (2000) A gated channel into the proteasome core particle. Nat Struct Mol Biol 7: 1062–1067 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous