The HLH-6 transcription factor regulates C. elegans pharyngeal gland development and function - PubMed (original) (raw)
The HLH-6 transcription factor regulates C. elegans pharyngeal gland development and function
Ryan B Smit et al. PLoS Genet. 2008 Oct.
Abstract
The Caenorhabditis elegans pharynx (or foregut) functions as a pump that draws in food (bacteria) from the environment. While the "organ identity factor" PHA-4 is critical for formation of the C. elegans pharynx as a whole, little is known about the specification of distinct cell types within the pharynx. Here, we use a combination of bioinformatics, molecular biology, and genetics to identify a helix-loop-helix transcription factor (HLH-6) as a critical regulator of pharyngeal gland development. HLH-6 is required for expression of a number of gland-specific genes, acting through a discrete cis-regulatory element named PGM1 (Pharyngeal Gland Motif 1). hlh-6 mutants exhibit a frequent loss of a subset of glands, while the remaining glands have impaired activity, indicating a role for hlh-6 in both gland development and function. Interestingly, hlh-6 mutants are also feeding defective, ascribing a biological function for the glands. Pharyngeal pumping in hlh-6 mutants is normal, but hlh-6 mutants lack expression of a class of mucin-related proteins that are normally secreted by pharyngeal glands and line the pharyngeal cuticle. An interesting possibility is that one function of pharyngeal glands is to secrete a pharyngeal lining that ensures efficient transport of food along the pharyngeal lumen.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
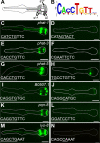
Figure 1. PGM1 is required for expression of some pharyngeal gland genes.
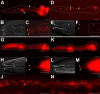
(A) Diagram of pharynx, highlighting the pharyngeal glands, modified from . (B) WebLogo of computationally identified PGM1. (C–N) Fluorescence micrographs of gland-expressed GFP or YFP reporters with wild-type promoter sequence (left column) or promoter sequence in which PGM1 is mutated (right column). In wild-type sequences (left) the E-box is underlined and in mutant sequences (right) the mutation is underlined. Anterior is at left and the pharynx is outlined. Scale bars represent 10 µm.
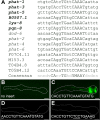
Figure 2. The extended PGM1 is sufficient for gland-specific expression.
(A) Alignment of PGM1 occurrences in the promoters of gland-expressed genes. Expression of genes in bold is experimentally verified to be both PGM1 and HLH-6 dependent. (B–E) Fluorescence micrographs of GFP enhancer constructs containing (B) no insert, (C) three tandem copies of the extended PGM1, (D) three tandem copies of the extended PGM1 in which the E-box has been mutated and (E) three tandem copies of the extended PGM1 in which sequence flanking the E-box has been altered. Anterior is at left and the pharynx is outlined. Scale bars represent 10 µm.
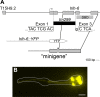
Figure 3. hlh-6 is expressed in pharyngeal glands.
(A) Schematic of the genomic region containing hlh-6. The position of the deletion allele tm299 is indicated. The portion of hlh-6 encoding the DNA Binding Domain (DBD) is shown as is the hlh-6 “minigene”, which rescues all aspects of the hlh-6 mutant phenotype. (B) Expression of the hlh-6::YFP reporter, containing 1175 bp (of 1190 bp) of intergenic sequence from the ATG of hlh-6 to just downstream of the stop codon of the next upstream gene, T15H9.2. Anterior is at left and the pharynx is outlined. Scale bar represents 10 µm.
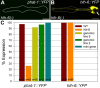
Figure 4. hlh-6 is required for PGM1 activity.
(A) An example of hlh-6 mutants where expression of phat-1::YFP is not visible. (B) An example of hlh-6 mutants where expression of hlh-6::YFP is visible only in g1 cells. The absence of the g2 cells is indicated by the arrow. Anterior is at left and the pharynx is outlined. Scale bars represent 10 µm. (C) Quantitation of the number of animals expressing each reporter in hlh-6 mutants. For the phat-1::YFP reporter in wildtype and hlh-6 mutants, only one transgenic line was scored but the same array was used in both genotypes. Two lines of the genomic rescue were scored for phat-1::YFP expression (lines 5 and 2). Only one line of minigene rescue was scored. Number of animals scored is indicated.
Figure 5. The g2 glands are not generated in hlh-6 mutants.
The lineages of the g2 glands in wild-type and hlh-6 mutants. MSn is used because both the MSa and MSp cell give rise to a g2 cell. If n = a, the g2L cell is made (as well as pm6VL and vpi2DL) and if n = p, the g2R cell is made (and pm6VR and vpi2DR). The sister cell of g2 cell undergoes apoptosis (X) in wild-type animals.
Figure 6. Phenotypic analysis of hlh-6 mutants.
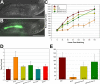
(A–B) The stuffed pharynx phenotype of hlh-6 mutants grown on OP50-GFP bacteria. (A) NDIC image, (B) merged NDIC and fluorescence image. Anterior is at left and scale bars represent 10 µm. (C–E) Assays for growth defects in wild-type, hlh-6 mutants and hlh-6 mutants rescued by either the hlh-6 genomic fragment, the hlh-6 minigene or by using the HB101 strain of E. coli. (C) Graph of body length versus time, (D) time to reach adulthood and (E) brood sizes. For the hlh-6 mutants the L1 arrested animals are omitted. Error bars represent one standard deviation.
Figure 7. Staining of intestinal fat stores of hlh-6 mutants.
(A–D) Fluorescence images of animals grown in the presence of Nile Red. (A) wild type, (B) daf-16(RNAi), (C) hlh-6 and (D) hlh-6; daf-16(RNAi). Anterior is at left and the pharynx is outlined. Scale bars represent 10 µm.
Figure 8. PHAT-5::MCHERRY localization in wild type and hlh-6 mutants.
Fluorescence and NDIC images of (A–C) wild-type and (D–F) hlh-6 animals expressing the hlh-6::phat-5::mCherry translational fusion construct. (B) and (C) are close-ups of animal shown in (A). (E) (F) are close-ups of (D). Fluorescence and NDIC images of (G) wild-type and (K) hlh-6 animals expressing the myo-2::phat-5::mCherry translational fusion construct with corresponding close-ups in (H–I) and (L–M). Arrowheads indicate the pharyngeal lumen, arrows mark the processes of the g1 glands and triangles mark the boundary of the pharyngeal cuticle. PHAT-5::MCHERRY is not found in the intestinal lumen of wild type animals (J) but is present in the intestinal lumen of hlh-6 mutants (N), indicated by carats. Anterior is at left and the pharynx is outlined. Scale bars represent 10 µm.
Similar articles
- Transcriptional regulation of HLH-6-independent and subtype-specific genes expressed in the Caenorhabditis elegans pharyngeal glands.
Ghai V, Smit RB, Gaudet J. Ghai V, et al. Mech Dev. 2012 Sep-Dec;129(9-12):284-97. doi: 10.1016/j.mod.2012.06.005. Epub 2012 Jul 1. Mech Dev. 2012. PMID: 22759833 - The CSL transcription factor LAG-1 directly represses hlh-6 expression in C. elegans.
Ghai V, Gaudet J. Ghai V, et al. Dev Biol. 2008 Oct 15;322(2):334-44. doi: 10.1016/j.ydbio.2008.07.018. Epub 2008 Jul 25. Dev Biol. 2008. PMID: 18706403 - Gland-specific expression of C. elegans hlh-6 requires the combinatorial action of three distinct promoter elements.
Raharjo I, Gaudet J. Raharjo I, et al. Dev Biol. 2007 Feb 1;302(1):295-308. doi: 10.1016/j.ydbio.2006.09.036. Epub 2006 Sep 26. Dev Biol. 2007. PMID: 17049341 - Development of Caenorhabditis elegans pharynx, with emphasis on its nervous system.
Pilon M, Mörck C. Pilon M, et al. Acta Pharmacol Sin. 2005 Apr;26(4):396-404. doi: 10.1111/j.1745-7254.2005.00070.x. Acta Pharmacol Sin. 2005. PMID: 15780187 Review. - The C. elegans pharynx: a model for organogenesis.
Mango SE. Mango SE. WormBook. 2007 Jan 22:1-26. doi: 10.1895/wormbook.1.129.1. WormBook. 2007. PMID: 18050503 Free PMC article. Review.
Cited by
- Extension of the Caenorhabditis elegans Pharyngeal M1 neuron axon is regulated by multiple mechanisms.
Refai O, Rohs P, Mains PE, Gaudet J. Refai O, et al. G3 (Bethesda). 2013 Nov 6;3(11):2015-29. doi: 10.1534/g3.113.008466. G3 (Bethesda). 2013. PMID: 24048649 Free PMC article. - C. elegans PEZO-1 is a mechanosensitive ion channel involved in food sensation.
Millet JRM, Romero LO, Lee J, Bell B, Vásquez V. Millet JRM, et al. J Gen Physiol. 2022 Jan 3;154(1):e202112960. doi: 10.1085/jgp.202112960. Epub 2021 Dec 2. J Gen Physiol. 2022. PMID: 34854875 Free PMC article. - Developmental genetics of the Caenorhabditis elegans pharynx.
Pilon M. Pilon M. Wiley Interdiscip Rev Dev Biol. 2014 Jul-Aug;3(4):263-80. doi: 10.1002/wdev.139. Epub 2014 May 23. Wiley Interdiscip Rev Dev Biol. 2014. PMID: 25262818 Free PMC article. Review. - Temporal Regulation of Gene Expression in Post-Mitotic Cells is Revealed from a Synchronized Population of C. elegans Larvae.
Roy PJ. Roy PJ. MicroPubl Biol. 2022 Jun 10;2022:10.17912/micropub.biology.000587. doi: 10.17912/micropub.biology.000587. eCollection 2022. MicroPubl Biol. 2022. PMID: 35783576 Free PMC article. - A lineage-resolved cartography of microRNA promoter activity in C. elegans empowers multidimensional developmental analysis.
Xu W, Liu J, Qi H, Si R, Zhao Z, Tao Z, Bai Y, Hu S, Sun X, Cong Y, Zhang H, Fan D, Xiao L, Wang Y, Li Y, Du Z. Xu W, et al. Nat Commun. 2024 Mar 30;15(1):2783. doi: 10.1038/s41467-024-47055-4. Nat Commun. 2024. PMID: 38555276 Free PMC article.
References
- Mango SE, Lambie EJ, Kimble J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development. 1994;120:3019–3031. - PubMed
- Kalb JM, Lau KK, Goszczynski B, Fukushige T, Moons D, et al. pha-4 is Ce-fkh-1, a fork head/HNF-3alpha,beta,gamma homolog that functions in organogenesis of the C. elegans pharynx. Development. 1998;125:2171–2180. - PubMed
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. - PubMed
- Roy Chowdhuri S, Crum T, Woollard A, Aslam S, Okkema PG. The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans. Dev Biol. 2006;295:664–677. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous