53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility - PubMed (original) (raw)
. 2008 Nov 27;456(7221):524-8.
doi: 10.1038/nature07433. Epub 2008 Oct 19.
Affiliations
- PMID: 18931659
- PMCID: PMC2613650
- DOI: 10.1038/nature07433
53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility
Nadya Dimitrova et al. Nature. 2008.
Abstract
Double-strand breaks activate the ataxia telangiectasia mutated (ATM) kinase, which promotes the accumulation of DNA damage factors in the chromatin surrounding the break. The functional significance of the resulting DNA damage foci is poorly understood. Here we show that 53BP1 (also known as TRP53BP1), a component of DNA damage foci, changes the dynamic behaviour of chromatin to promote DNA repair. We used conditional deletion of the shelterin component TRF2 (also known as TERF2) from mouse cells (TRF2(fl/-)) to deprotect telomeres, which, like double-strand breaks, activate the ATM kinase, accumulate 53BP1 and are processed by non-homologous end joining (NHEJ). Deletion of TRF2 from 53BP1-deficient cells established that NHEJ of dysfunctional telomeres is strongly dependent on the binding of 53BP1 to damaged chromosome ends. To address the mechanism by which 53BP1 promotes NHEJ, we used time-lapse microscopy to measure telomere dynamics before and after their deprotection. Imaging showed that deprotected telomeres are more mobile and sample larger territories within the nucleus. This change in chromatin dynamics was dependent on 53BP1 and ATM but did not require a functional NHEJ pathway. We propose that the binding of 53BP1 near DNA breaks changes the dynamic behaviour of the local chromatin, thereby facilitating NHEJ repair reactions that involve distant sites, including joining of dysfunctional telomeres and AID (also known as AICDA)-induced breaks in immunoglobulin class-switch recombination.
Figures
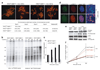
Figure 1. Requirement for 53BP1 in NHEJ of dysfunctional telomeres, but not DNA damage signaling
a, (Top) Metaphase chromosomes after deletion of TRF2 from the indicated cells. Telomeric FISH, green; DAPI, red. (Bottom) Summary of the effect of 53BP1 on telomere fusions at 120 hours after TRF2 deletion. b, In-gel assay for the 3’ overhang (left) and total telomeric DNA (right) after deletion of TRF2 from 53BP1-proficient and -deficient cells. c, Quantification of overhang signals in (b) (mean±SD; n=3). d, IF for presence of γ-H2AX at telomeres after deletion of TRF2 from 53BP1-proficent and -deficient cells. e, Immunoblots for phosphorylated ATM and Chk2 after TRF2 deletion. f, Proliferation of MEFs of the indicated genotype and treatment (mean±SD; n=3).
Figure 2. Optimal NHEJ of dysfunctional telomeres requires interaction of 53BP1 with H4-K20diMe
a, Schematic of the domain structure of 53BP1. b, Immunoblot for 53BP1 expression in TRF2F/−53BP1−/− cells complemented with empty vector, wild type (WT), or the Tudor mutant 53BP1-D1521A. c, Telomere fusions in metaphase spreads of TRF2F/−53BP1−/− MEFs complemented as indicated and treated with Cre as specified. d, Quantification of telomere fusions in (c) (mean±SD; n=3).
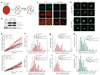
Figure 3. 53BP1 promotes mobility of dysfunctional telomeres
a, Schematic of the live-cell imaging experiments. b, Immunoblotting for EGFP-TRF1, mCherry-BP1-2, and TRF2. c, Co-localization of EGFP-TRF1 and mCherry-BP1-2 fluorescence signals. d, Representative traces of telomeres in indicated MEFs expressing EGFP-TRF1 and mCherry-BP1-2. Snapshots taken at indicated timepoints. e, 10 representative telomeres from indicated MEFs, treated as specified, were tracked as in (d); cumulative distance at each time-point was plotted against time. f, Frequency distribution of the cumulative distance traveled in one representative experiment in MEFs of the indicated genotype and treatment (median±SD.; n=55; p calculated using a two-tailed Mann-Whitney test). g, Frequency distribution of the maximum displacement from the starting point registered by individual telomeres during the imaging session described in (f). (median±SD and p-values calculated as in (f)). h, i, Analysis as in (f) in MEFs of the indicated genotypes and treatments, expressing EGFP-TRF1 marker only (n=50).
Similar articles
- DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion.
Celli GB, de Lange T. Celli GB, et al. Nat Cell Biol. 2005 Jul;7(7):712-8. doi: 10.1038/ncb1275. Epub 2005 Jun 19. Nat Cell Biol. 2005. PMID: 15968270 - Multiple roles for MRE11 at uncapped telomeres.
Deng Y, Guo X, Ferguson DO, Chang S. Deng Y, et al. Nature. 2009 Aug 13;460(7257):914-8. doi: 10.1038/nature08196. Epub 2009 Jul 26. Nature. 2009. PMID: 19633651 Free PMC article. - DNA-damage response and repair activities at uncapped telomeres depend on RNF8.
Peuscher MH, Jacobs JJ. Peuscher MH, et al. Nat Cell Biol. 2011 Aug 21;13(9):1139-45. doi: 10.1038/ncb2326. Nat Cell Biol. 2011. PMID: 21857671 - Double-strand break repair: 53BP1 comes into focus.
Panier S, Boulton SJ. Panier S, et al. Nat Rev Mol Cell Biol. 2014 Jan;15(1):7-18. doi: 10.1038/nrm3719. Epub 2013 Dec 11. Nat Rev Mol Cell Biol. 2014. PMID: 24326623 Review. - The Connection Between Cell Fate and Telomere.
Engin AB, Engin A. Engin AB, et al. Adv Exp Med Biol. 2021;1275:71-100. doi: 10.1007/978-3-030-49844-3_3. Adv Exp Med Biol. 2021. PMID: 33539012 Review.
Cited by
- DNA repair at telomeres: keeping the ends intact.
Webb CJ, Wu Y, Zakian VA. Webb CJ, et al. Cold Spring Harb Perspect Biol. 2013 Jun 1;5(6):a012666. doi: 10.1101/cshperspect.a012666. Cold Spring Harb Perspect Biol. 2013. PMID: 23732473 Free PMC article. Review. - Put a RING on it: regulation and inhibition of RNF8 and RNF168 RING finger E3 ligases at DNA damage sites.
Bartocci C, Denchi EL. Bartocci C, et al. Front Genet. 2013 Jul 9;4:128. doi: 10.3389/fgene.2013.00128. eCollection 2013. Front Genet. 2013. PMID: 23847653 Free PMC article. - Cas9 deactivation with photocleavable guide RNAs.
Zou RS, Liu Y, Wu B, Ha T. Zou RS, et al. Mol Cell. 2021 Apr 1;81(7):1553-1565.e8. doi: 10.1016/j.molcel.2021.02.007. Epub 2021 Mar 3. Mol Cell. 2021. PMID: 33662274 Free PMC article. - RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection.
Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ. Chapman JR, et al. Mol Cell. 2013 Mar 7;49(5):858-71. doi: 10.1016/j.molcel.2013.01.002. Epub 2013 Jan 17. Mol Cell. 2013. PMID: 23333305 Free PMC article. - 53BP1 promotes microhomology-mediated end-joining in G1-phase cells.
Xiong X, Du Z, Wang Y, Feng Z, Fan P, Yan C, Willers H, Zhang J. Xiong X, et al. Nucleic Acids Res. 2015 Feb 18;43(3):1659-70. doi: 10.1093/nar/gku1406. Epub 2015 Jan 13. Nucleic Acids Res. 2015. PMID: 25586219 Free PMC article.
References
- Celli G, de Lange T. DNA processing not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. - PubMed
- Celli GB, Lazzerini Denchi E, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885–890. - PubMed
- Lazzerini Denchi E, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. - PubMed
- DiTullio RA, Jr, et al. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol. 2002;4:998–1002. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- EY18244/EY/NEI NIH HHS/United States
- OD000379/OD/NIH HHS/United States
- R01 GM049046/GM/NIGMS NIH HHS/United States
- DP1 OD000379-04/OD/NIH HHS/United States
- GM42694/GM/NIGMS NIH HHS/United States
- R37 GM049046/GM/NIGMS NIH HHS/United States
- DP1 OD000379/OD/NIH HHS/United States
- PN2 EY018244/EY/NEI NIH HHS/United States
- Howard Hughes Medical Institute/United States
- GM049046/GM/NIGMS NIH HHS/United States
- R01 GM042694/GM/NIGMS NIH HHS/United States
- R37 GM049046-16/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous