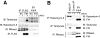Proteomics-based identification of novel factor inhibiting hypoxia-inducible factor (FIH) substrates indicates widespread asparaginyl hydroxylation of ankyrin repeat domain-containing proteins - PubMed (original) (raw)
Proteomics-based identification of novel factor inhibiting hypoxia-inducible factor (FIH) substrates indicates widespread asparaginyl hydroxylation of ankyrin repeat domain-containing proteins
Matthew E Cockman et al. Mol Cell Proteomics. 2009 Mar.
Abstract
Post-translational hydroxylation has been considered an unusual modification on intracellular proteins. However, following the recognition that oxygen-sensitive prolyl and asparaginyl hydroxylation are central to the regulation of the transcription factor hypoxia-inducible factor (HIF), interest has centered on the possibility that these enzymes may have other substrates in the proteome. In support of this certain ankyrin repeat domain (ARD)-containing proteins, including members of the IkappaB and Notch families, have been identified as alternative substrates of the HIF asparaginyl hydroxylase factor inhibiting HIF (FIH). Although these findings imply a potentially broad range of substrates for FIH, the precise extent of this range has been difficult to determine because of the difficulty of capturing transient enzyme-substrate interactions. Here we describe the use of pharmacological "substrate trapping" together with stable isotope labeling by amino acids in cell culture (SILAC) technology to stabilize and identify potential FIH-substrate interactions by mass spectrometry. To pursue these potential FIH substrates we used conventional data-directed tandem MS together with alternating low/high collision energy tandem MS to assign and quantitate hydroxylation at target asparaginyl residues. Overall the work has defined 13 new FIH-dependent hydroxylation sites with a degenerate consensus corresponding to that of the ankyrin repeat and a range of ARD-containing proteins as actual and potential substrates for FIH. Several ARD-containing proteins were multiply hydroxylated, and detailed studies of one, Tankyrase-2, revealed eight sites that were differentially sensitive to FIH-catalyzed hydroxylation. These findings indicate that asparaginyl hydroxylation is likely to be widespread among the approximately 300 ARD-containing species in the human proteome.
Figures
Fig. 1.
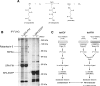
Substrate trapping with DMOG identifies ARD proteins as novel FIH interactors. A, reaction scheme for FIH-dependent hydroxylation and its inhibition by 2OG analogues. FIH-mediated hydroxylation of substrate (R) is coupled to the oxidative decarboxylation of 2OG in a reaction that consumes molecular oxygen and that generates succinate and CO2. Analogues of 2OG, _e.g. N_-oxalylglycine (and the cell-penetrant diester form DMOG) in which a secondary amine has been substituted for a methylene group at the 2OG 3-position, competitively inhibit FIH activity. B, identification of ARD-containing proteins by affinity purification/SDS-PAGE. Coomassie Blue stain of preparative anti-FLAG immunoprecipitations from HEK293 cells stably expressing SPA-tagged FIH (SPA tandem affinity tag: 3× FLAG tag-tobacco etch virus cleavage site-calmodulin binding peptide) or SPA-tagged EGFP (control) is shown. Addition of 1 m
m
DMOG for 16 h stabilized interactions with two proteins of ∼95 and 130 kDa in an FIH-dependent manner. Tandem MS of tryptic digests assigned the 95-kDa species as RIPK4 and the 130-kDa species as Rabankyrin-5. C, synopsis of the SILAC work flow. Two isotopically distinct populations of FLAG-tagged FIH-inducible U2OS cells (tet-FIH) were prepared by passaging cells in either normal medium (Lys0/Arg0) or medium containing heavy isotopes (Lys6/Arg10) for 7 days using standard SILAC procedures. Both populations were treated with doxycycline (Dox) for 18 h (to induce FIH expression), but only the heavy population of cells was exposed to 1 m
m
DMOG for 16 h. Lysates were prepared, and FIH complexes were immunopurified using FLAG affinity agarose. The agarose gel was washed, pooled, and eluted before desalting and digestion with trypsin. To improve the assignment of peptides, MS/MS data were collected on a Q-TOF tandem mass spectrometer in both DDA and data-independent (MSE) modes. An increased ratio of heavy (H) to light (L) peptides (i.e. above the ratio observed for the bait peptides) suggested that the protein was binding in a DMOG-sensitive and substrate-specific way. To distinguish between constitutive interactions and contaminants in the SILAC screen, a second control (FLAG) IP was performed in mock transfected cells (tet-EV) grown in normal medium supplemented with doxycycline and DMOG. In-solution digests of material eluted from the control IP were subjected to identical MS/MS analyses and compared against the protein lists generated from the SILAC experiment.
Fig. 2.
Interactions between FIH and Rabankyrin-5, RNase L, and Tankyrase-2 in cells. A, ARD-containing proteins interact with FIH. FLAG-tagged FIH-inducible U2OS cells (FIH) or the parental line (EV) were treated with 0.5 μg/ml doxycycline in the presence or absence of DMOG for 18 h prior to FLAG IP and anti-FLAG or anti-Rabankyrin-5/RNase L/Tankyrase immunoblotting (IB). For all putative substrates, addition of 1 m
m
DMOG for 16 h stabilized FIH-ARD interactions. B, co-immunoprecipitation of endogenous FIH with RNase L, Rabankyrin-5, and Tankyrase. Anti-ARD IPs (anti-Rabankyrin, anti-Tankyrase, and anti-RNase L) and not control IPs (species/isotype-matched control IgG) co-precipitate endogenous FIH in U2OS cells. For all putative substrates, addition of 1 m
m
DMOG to cells for 16 h stabilized FIH-ARD complexes.
Fig. 3.
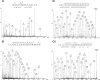
RNase L, Rabankyrin, and Tankyrase-2 are hydroxylated in vivo. MS/MS assignment of asparaginyl hydroxylation in ARD substrates that were immunopurified from transfected 293T cells under physiological levels of FIH is shown. A, MS/MS of the tryptic peptide HGAVVNVADLWK ([M + 2H]2+ = m/z 662.86) in Tankyrase-2 assigns hydroxylation at Asn-586; a mass identical to a hydroxyasparagine-containing fragment is observed in the b ion series at the b6 ion. Where present, y0 indicates a y ion with a loss of a water molecule (for Asn-706 and Asn-739 spectra, see supplemental Fig. S1, A and B, respectively). B, MS/MS spectra of the tryptic peptide NALIHALLSSDDSDVEAITHLLLDHGADVNVR ([M + 4H]4+ = m/z 860.70) in RNase L assigns hydroxylation at Asn-233; a mass identical to a hydroxyasparagine-containing fragment is observed in the y ion series at the y3 ion. C, MS/MS spectra of the tryptic peptide DGQTPLHLAASWGLEETVQCLLEFGANVNAQDAEGR derived from Rabankyrin-5 in the non-hydroxylated m/z 1299.64 ([M + 3H]3) (i) and hydroxylated m/z 1304.97 ([M + 3H]3+) state (ii); a +16-Da shift appears in the b and y ion series of ii at b29 and y8, corresponding to fragments containing Asn-797. Both peptides carry a carbamidomethylated cysteine modification.
Fig. 4.
Quantitative mass spectrometric analysis of Tankyrase-2 hydroxylation and assignment by data-independent MS/MS (MSE). Parallel MS/MS assignment and quantitation of Asn-739 hydroxylation in Tankyrase-2 that was immunopurified from transiently transfected 293T cells are shown. A, MS/MS (MSE) spectra of the parental m/z 578.26 ([M + 2H]2+) (i) and hydroxylated m/z 586.26 ([M + 2H]2+) (ii) YNACVNATDK tryptic peptide; a +16-Da shift is observed at the y5 ion corresponding to fragments containing Asn-739 (see supplemental Fig. S1B for unambiguous assignment of Asn-739 hydroxylation by DDA MS/MS). Both parent and hydroxylated ions are carbamidomethylated cysteine-modified. B, extracted ion chromatograms for parent and hydroxylated YNACVNATDK ions m/z 578.26 and m/z 586.26, respectively. Data derived from UPLC-MSE chromatography runs illustrating the relative abundance of hydroxylation at Asn-739 (46% OH) under endogenous levels of FIH are shown.
Fig. 5.
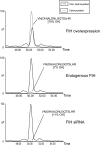
FIH-dependent hydroxylation at Asn-427 in Tankyrase-2. Extracted ion chromatograms of m/z 513.27 and m/z 518.60 corresponding to unmodified and hydroxylated forms of the tryptic Tankyrase-2 peptide VNALDNLGQTSLHR ([M + 3H]3+). Nano-UPLC-MSE chromatography analysis illustrating the relative abundance of hydroxylation at Asn-427 following FIH intervention: FIH RNA interference knockdown (lower panel; <1%), endogenous levels of FIH (middle panel; ∼7%), and FIH overexpression (upper panel; ∼70%) is shown.
Fig. 6.
Distribution of amino acids surrounding all known sites of FIH-dependent asparaginyl hydroxylation. A, ClustalW non-gapped multiple sequence alignment of all known FIH-mediated hydroxylation sites. Novel sites identified in Tankyrase-2, RNase L, and Rabankyrin-5 are indicated. B, a revised FIH hydroxylation consensus. Logo representation of non-gapped ClustalW alignment displaying all known asparaginyl hydroxylation sites in comparison with the AR consensus (23) is shown. Residues homologous (>25% frequency in the FIH consensus) to the AR consensus are indicated (+). Sequence analysis was carried out using the Logo server (WebLogo).
Comment in
- Substrates of PHD.
Lee FS. Lee FS. Cell Metab. 2019 Oct 1;30(4):626-627. doi: 10.1016/j.cmet.2019.08.008. Cell Metab. 2019. PMID: 31577931 Free PMC article.
Similar articles
- MYPT1, the targeting subunit of smooth-muscle myosin phosphatase, is a substrate for the asparaginyl hydroxylase factor inhibiting hypoxia-inducible factor (FIH).
Webb JD, Murányi A, Pugh CW, Ratcliffe PJ, Coleman ML. Webb JD, et al. Biochem J. 2009 May 13;420(2):327-33. doi: 10.1042/BJ20081905. Biochem J. 2009. PMID: 19245366 - Posttranslational hydroxylation of ankyrin repeats in IkappaB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH).
Cockman ME, Lancaster DE, Stolze IP, Hewitson KS, McDonough MA, Coleman ML, Coles CH, Yu X, Hay RT, Ley SC, Pugh CW, Oldham NJ, Masson N, Schofield CJ, Ratcliffe PJ. Cockman ME, et al. Proc Natl Acad Sci U S A. 2006 Oct 3;103(40):14767-72. doi: 10.1073/pnas.0606877103. Epub 2006 Sep 26. Proc Natl Acad Sci U S A. 2006. PMID: 17003112 Free PMC article. - Asparaginyl beta-hydroxylation of proteins containing ankyrin repeat domains influences their stability and function.
Hardy AP, Prokes I, Kelly L, Campbell ID, Schofield CJ. Hardy AP, et al. J Mol Biol. 2009 Oct 2;392(4):994-1006. doi: 10.1016/j.jmb.2009.07.070. Epub 2009 Jul 30. J Mol Biol. 2009. PMID: 19646994 - FIH-dependent asparaginyl hydroxylation of ankyrin repeat domain-containing proteins.
Cockman ME, Webb JD, Ratcliffe PJ. Cockman ME, et al. Ann N Y Acad Sci. 2009 Oct;1177:9-18. doi: 10.1111/j.1749-6632.2009.05042.x. Ann N Y Acad Sci. 2009. PMID: 19845602 Review. - Signalling cross talk of the HIF system: involvement of the FIH protein.
Coleman ML, Ratcliffe PJ. Coleman ML, et al. Curr Pharm Des. 2009;15(33):3904-7. doi: 10.2174/138161209789649448. Curr Pharm Des. 2009. PMID: 19671041 Review.
Cited by
- Asparagine Hydroxylation is a Reversible Post-translational Modification.
Rodriguez J, Haydinger CD, Peet DJ, Nguyen LK, von Kriegsheim A. Rodriguez J, et al. Mol Cell Proteomics. 2020 Nov;19(11):1777-1789. doi: 10.1074/mcp.RA120.002189. Epub 2020 Aug 5. Mol Cell Proteomics. 2020. PMID: 32759169 Free PMC article. - Prolyl hydroxylase substrate adenylosuccinate lyase is an oncogenic driver in triple negative breast cancer.
Zurlo G, Liu X, Takada M, Fan C, Simon JM, Ptacek TS, Rodriguez J, von Kriegsheim A, Liu J, Locasale JW, Robinson A, Zhang J, Holler JM, Kim B, Zikánová M, Bierau J, Xie L, Chen X, Li M, Perou CM, Zhang Q. Zurlo G, et al. Nat Commun. 2019 Nov 15;10(1):5177. doi: 10.1038/s41467-019-13168-4. Nat Commun. 2019. PMID: 31729379 Free PMC article. - PHD3 Regulates p53 Protein Stability by Hydroxylating Proline 359.
Rodriguez J, Herrero A, Li S, Rauch N, Quintanilla A, Wynne K, Krstic A, Acosta JC, Taylor C, Schlisio S, von Kriegsheim A. Rodriguez J, et al. Cell Rep. 2018 Jul 31;24(5):1316-1329. doi: 10.1016/j.celrep.2018.06.108. Cell Rep. 2018. PMID: 30067985 Free PMC article. - Decoding the proteomic changes involved in the biofilm formation of Enterococcus faecalis SK460 to elucidate potential biofilm determinants.
Suryaletha K, Narendrakumar L, John J, Radhakrishnan MP, George S, Thomas S. Suryaletha K, et al. BMC Microbiol. 2019 Jun 28;19(1):146. doi: 10.1186/s12866-019-1527-2. BMC Microbiol. 2019. PMID: 31253082 Free PMC article. - Factor inhibiting HIF (FIH-1) promotes renal cancer cell survival by protecting cells from HIF-1α-mediated apoptosis.
Khan MN, Bhattacharyya T, Andrikopoulos P, Esteban MA, Barod R, Connor T, Ashcroft M, Maxwell PH, Kiriakidis S. Khan MN, et al. Br J Cancer. 2011 Mar 29;104(7):1151-9. doi: 10.1038/bjc.2011.73. Epub 2011 Mar 8. Br J Cancer. 2011. PMID: 21386837 Free PMC article.
References
- Walsh, C. T. ( 2006) Post Translational Modification of Proteins: Expanding Nature's Inventory, pp. 331–347, Roberts and Co. Publishers, Greenwood Village, CO
- Schofield, C. J., and Ratcliffe, P. J. ( 2004) Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 5, 343–354 - PubMed
- Cockman, M. E., Lancaster, D. E., Stolze, I. P., Hewitson, K. S., McDonough, M. A., Coleman, M. L., Coles, C. H., Yu, X., Hay, R. T., Ley, S. C., Pugh, C. W., Oldham, N. J., Masson, N., Schofield, C. J., and Ratcliffe, P. J. ( 2006) Posttranslational hydroxylation of ankyrin repeats in IκB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH). Proc. Natl. Acad. Sci. U. S. A. 103, 14767–14772 - PMC - PubMed
- Coleman, M. L., McDonough, M. A., Hewitson, K. S., Coles, C., Mecinovic, J., Edelmann, M., Cook, K. M., Cockman, M. E., Lancaster, D. E., Kessler, B. M., Oldham, N. J., Ratcliffe, P. J., and Schofield, C. J. ( 2007) Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J. Biol. Chem. 282, 24027–24038 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases